You are browsing environment: FUNGIDB
I302_08034-t42_1-p1
Basic Information
help
Species
Kwoniella bestiolae
Lineage
Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella bestiolae
CAZyme ID
I302_08034-t42_1-p1
CAZy Family
GT33
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
428
KI894025|CGC5 45106.39
7.2455
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_KbestiolaeCBS10118
9270
1296100
137
9133
Gene Location
No EC number prediction in I302_08034-t42_1-p1.
Family
Start
End
Evalue
family coverage
PL14
219
424
1.2e-96
0.9907834101382489
I302_08034-t42_1-p1 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.60e-129
178
428
249
499
9.10e-129
178
428
249
499
9.10e-129
178
428
249
499
4.85e-128
178
428
247
497
1.66e-127
178
428
253
503
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
1.34e-27
261
428
97
274
Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai]
more
I302_08034-t42_1-p1 has no Swissprot hit.
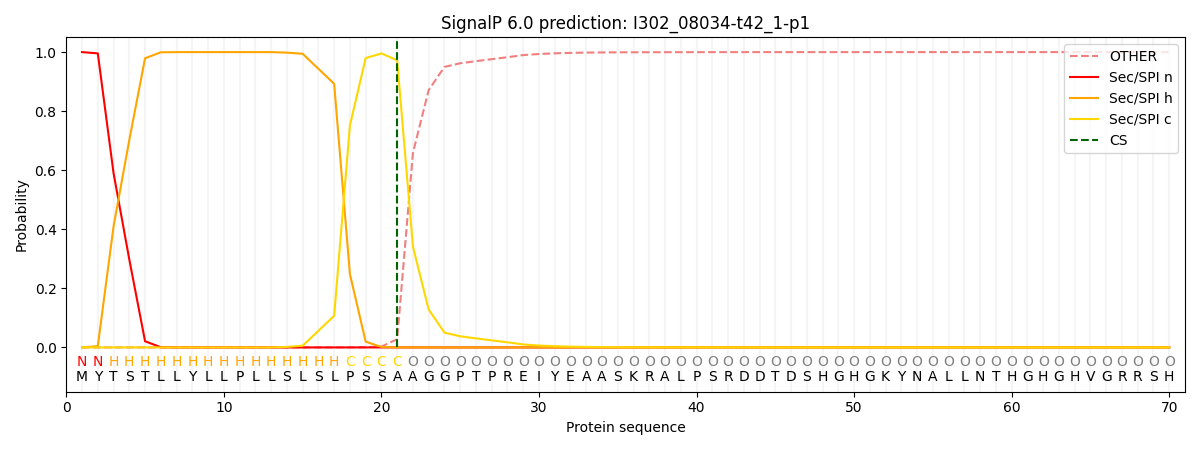
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000298
0.999680
CS pos: 21-22. Pr: 0.9721
There is no transmembrane helices in I302_08034-t42_1-p1.