You are browsing environment: FUNGIDB
CAZyme Information: I302_07725-t42_1-p1
You are here: Home > Sequence: I302_07725-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kwoniella bestiolae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella bestiolae | |||||||||||
| CAZyme ID | I302_07725-t42_1-p1 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.-:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA14 | 20 | 297 | 1.8e-85 | 0.9882352941176471 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396406 | WSC | 3.24e-19 | 615 | 692 | 2 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 396406 | WSC | 8.43e-19 | 433 | 509 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 396406 | WSC | 1.78e-18 | 322 | 400 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 214616 | WSC | 2.05e-15 | 318 | 410 | 1 | 95 | present in yeast cell wall integrity and stress response component proteins. Domain present in WSC proteins, polycystin and fungal exoglucanase |
| 214616 | WSC | 1.30e-13 | 429 | 517 | 1 | 93 | present in yeast cell wall integrity and stress response component proteins. Domain present in WSC proteins, polycystin and fungal exoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.05e-75 | 17 | 702 | 21 | 604 | |
| 5.74e-73 | 9 | 296 | 15 | 295 | |
| 1.07e-71 | 3 | 295 | 1 | 278 | |
| 3.24e-54 | 18 | 298 | 18 | 278 | |
| 1.36e-53 | 75 | 430 | 12 | 347 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.18e-49 | 20 | 296 | 1 | 261 | Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina],5NO7_B Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina] |
|
| 1.90e-10 | 322 | 407 | 91 | 178 | Wnt modulator Kremen in complex with DKK1 (CRD2) and LRP6 (PE3PE4) [Homo sapiens] |
|
| 3.42e-10 | 322 | 407 | 100 | 187 | Chain E, KRM1 [Homo sapiens],7BZU_E Chain E, KRM1 [Homo sapiens] |
|
| 3.47e-10 | 322 | 407 | 103 | 190 | Chain E, Kremen protein 1 [Homo sapiens] |
|
| 3.93e-10 | 322 | 407 | 131 | 218 | Wnt modulator Kremen crystal form I at 1.90A [Homo sapiens],5FWT_A Wnt modulator Kremen crystal form I at 2.10A [Homo sapiens],5FWU_A Wnt modulator Kremen crystal form II at 2.8A [Homo sapiens],5FWV_A Wnt modulator Kremen crystal form III at 3.2A [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.39e-33 | 319 | 700 | 63 | 402 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
|
| 1.65e-23 | 241 | 516 | 443 | 715 | WSC domain-containing protein ARB_07870 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07870 PE=1 SV=1 |
|
| 3.26e-23 | 332 | 516 | 29 | 219 | Putative fungistatic metabolite OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=CHGG_05463 PE=1 SV=2 |
|
| 1.65e-16 | 322 | 408 | 131 | 217 | WSC domain-containing protein 2 OS=Homo sapiens OX=9606 GN=WSCD2 PE=2 SV=2 |
|
| 1.68e-16 | 322 | 408 | 137 | 223 | WSC domain-containing protein 2 OS=Mus musculus OX=10090 GN=Wscd2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
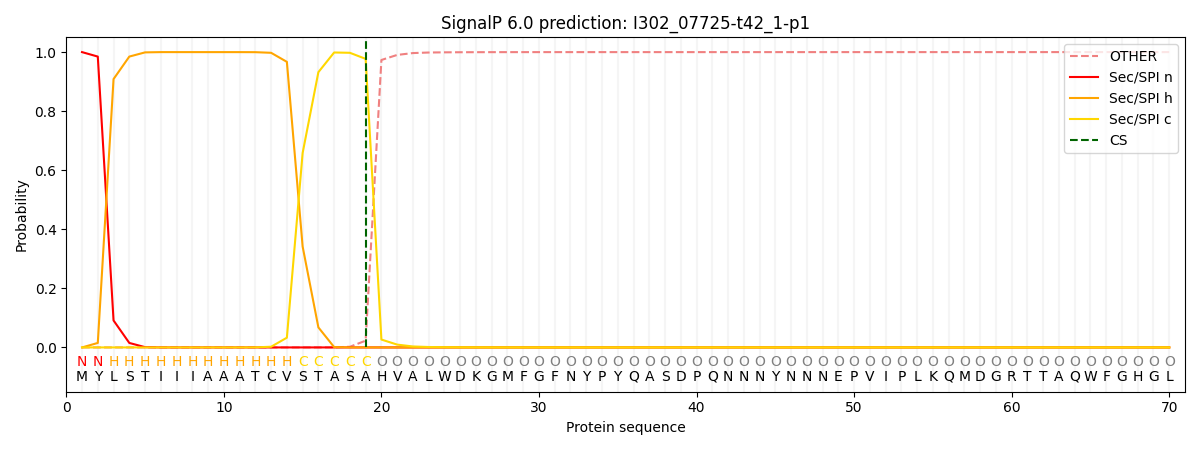
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000291 | 0.999681 | CS pos: 19-20. Pr: 0.9771 |

