You are browsing environment: FUNGIDB
CAZyme Information: I302_06222-t42_1-p1
You are here: Home > Sequence: I302_06222-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
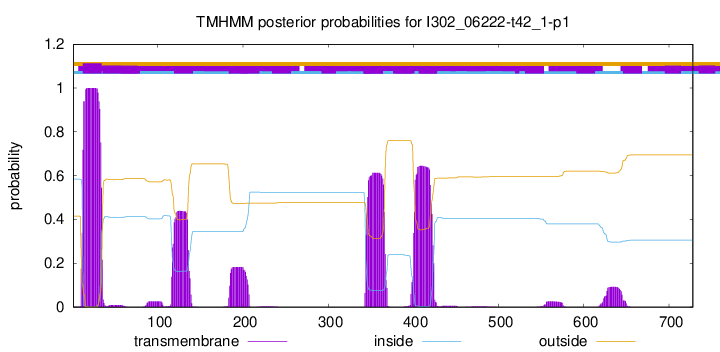
TMHMM annotations
Basic Information help
| Species | Kwoniella bestiolae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella bestiolae | |||||||||||
| CAZyme ID | I302_06222-t42_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | phosphatidylinositol glycan, class A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.198:3 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340827 | GT4_PIG-A-like | 0.0 | 345 | 728 | 1 | 382 | phosphatidylinositol N-acetylglucosaminyltransferase subunit A and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. Phosphatidylinositol glycan-class A (PIG-A), an X-linked gene in humans, is necessary for the synthesis of N-acetylglucosaminyl-phosphatidylinositol, a very early intermediate in glycosyl phosphatidylinositol (GPI)-anchor biosynthesis. The GPI-anchor is an important cellular structure that facilitates the attachment of many proteins to cell surfaces. Somatic mutations in PIG-A have been associated with Paroxysmal Nocturnal Hemoglobinuria (PNH), an acquired hematological disorder. |
| 340831 | GT4_PimA-like | 9.16e-57 | 346 | 712 | 2 | 366 | phosphatidyl-myo-inositol mannosyltransferase. This family is most closely related to the GT4 family of glycosyltransferases and named after PimA in Propionibacterium freudenreichii, which is involved in the biosynthesis of phosphatidyl-myo-inositol mannosides (PIM) which are early precursors in the biosynthesis of lipomannans (LM) and lipoarabinomannans (LAM), and catalyzes the addition of a mannosyl residue from GDP-D-mannose (GDP-Man) to the position 2 of the carrier lipid phosphatidyl-myo-inositol (PI) to generate a phosphatidyl-myo-inositol bearing an alpha-1,2-linked mannose residue (PIM1). Glycosyltransferases catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. The acceptor molecule can be a lipid, a protein, a heterocyclic compound, or another carbohydrate residue. This group of glycosyltransferases is most closely related to the previously defined glycosyltransferase family 1 (GT1). The members of this family may transfer UDP, ADP, GDP, or CMP linked sugars. The diverse enzymatic activities among members of this family reflect a wide range of biological functions. The protein structure available for this family has the GTB topology, one of the two protein topologies observed for nucleotide-sugar-dependent glycosyltransferases. GTB proteins have distinct N- and C- terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. The members of this family are found mainly in certain bacteria and archaea. |
| 153251 | LPLAT_AGPAT-like | 4.66e-51 | 55 | 244 | 7 | 184 | Lysophospholipid Acyltransferases (LPLATs) of Glycerophospholipid Biosynthesis: AGPAT-like. Lysophospholipid acyltransferase (LPLAT) superfamily member: acyltransferases of de novo and remodeling pathways of glycerophospholipid biosynthesis which catalyze the incorporation of an acyl group from either acylCoAs or acyl-acyl carrier proteins (acylACPs) into acceptors such as glycerol 3-phosphate, dihydroxyacetone phosphate or lyso-phosphatidic acid. Included in this subgroup are such LPLATs as 1-acyl-sn-glycerol-3-phosphate acyltransferase (AGPAT, PlsC), Tafazzin (product of Barth syndrome gene), and similar proteins. |
| 223515 | RfaB | 3.12e-46 | 346 | 716 | 3 | 379 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| 223282 | PlsC | 3.28e-41 | 26 | 251 | 18 | 227 | 1-acyl-sn-glycerol-3-phosphate acyltransferase [Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 728 | 1 | 735 | |
| 0.0 | 1 | 728 | 1 | 727 | |
| 0.0 | 1 | 728 | 1 | 727 | |
| 0.0 | 1 | 728 | 1 | 727 | |
| 0.0 | 1 | 667 | 1 | 656 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.83e-23 | 82 | 235 | 74 | 224 | Crystal Structure of the 1-acyl-sn-glycerophosphate (LPA) acyltransferase, PlsC, from Thermotoga maritima [Thermotoga maritima MSB8],5KYM_B Crystal Structure of the 1-acyl-sn-glycerophosphate (LPA) acyltransferase, PlsC, from Thermotoga maritima [Thermotoga maritima MSB8] |
|
| 7.65e-14 | 344 | 711 | 15 | 396 | Structure of Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM [Mycolicibacterium smegmatis MC2 155],6TVP_B Structure of Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM [Mycolicibacterium smegmatis MC2 155] |
|
| 2.02e-09 | 344 | 712 | 5 | 365 | Crystal structure of phosphatidyl mannosyltransferase PimA [Mycolicibacterium smegmatis MC2 155],4NC9_A Crystal structure of phosphatidyl mannosyltransferase PimA [Mycolicibacterium smegmatis MC2 155],4NC9_B Crystal structure of phosphatidyl mannosyltransferase PimA [Mycolicibacterium smegmatis MC2 155],4NC9_C Crystal structure of phosphatidyl mannosyltransferase PimA [Mycolicibacterium smegmatis MC2 155],4NC9_D Crystal structure of phosphatidyl mannosyltransferase PimA [Mycolicibacterium smegmatis MC2 155] |
|
| 2.15e-09 | 344 | 712 | 21 | 381 | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man [Mycolicibacterium smegmatis MC2 155],2GEK_A Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP [Mycolicibacterium smegmatis MC2 155] |
|
| 3.37e-09 | 355 | 681 | 13 | 338 | Chain A, Glycosyltransferase [Staphylococcus aureus subsp. aureus CN1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.18e-137 | 348 | 712 | 1 | 363 | Phosphatidylinositol N-acetylglucosaminyltransferase gpi3 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gpi3 PE=3 SV=1 |
|
| 1.40e-125 | 345 | 728 | 34 | 416 | Phosphatidylinositol N-acetylglucosaminyltransferase subunit A OS=Mus musculus OX=10090 GN=Piga PE=2 SV=1 |
|
| 1.91e-125 | 345 | 713 | 34 | 400 | Phosphatidylinositol N-acetylglucosaminyltransferase subunit A OS=Homo sapiens OX=9606 GN=PIGA PE=1 SV=1 |
|
| 3.59e-124 | 343 | 728 | 6 | 387 | Phosphatidylinositol N-acetylglucosaminyltransferase subunit A OS=Arabidopsis thaliana OX=3702 GN=PIGA PE=2 SV=1 |
|
| 3.54e-108 | 344 | 719 | 3 | 383 | Phosphatidylinositol N-acetylglucosaminyltransferase GPI3 subunit OS=Saccharomyces cerevisiae (strain RM11-1a) OX=285006 GN=SPT14 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
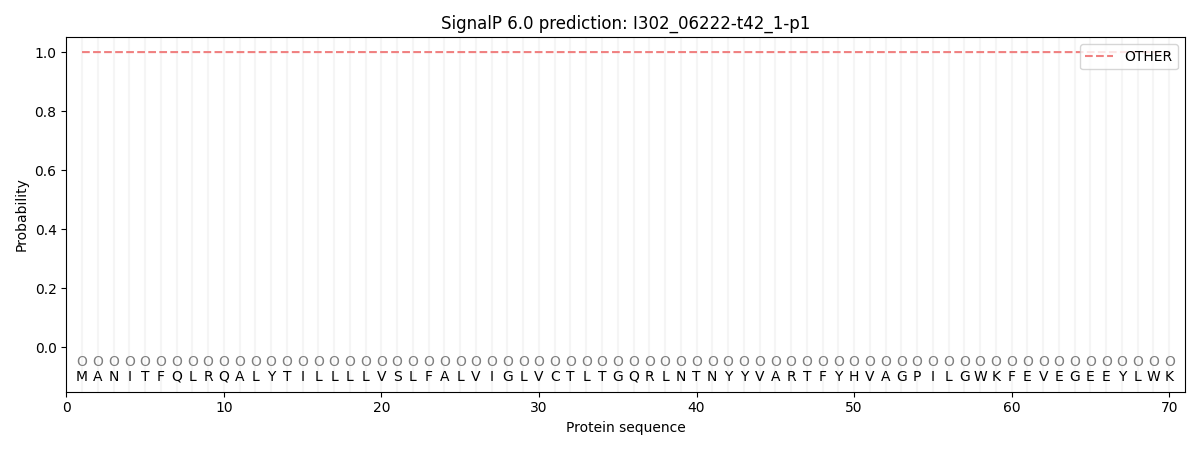
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000040 | 0.000001 |