You are browsing environment: FUNGIDB
CAZyme Information: I302_01862-t42_1-p1
You are here: Home > Sequence: I302_01862-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kwoniella bestiolae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella bestiolae | |||||||||||
| CAZyme ID | I302_01862-t42_1-p1 | |||||||||||
| CAZy Family | GH1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:20 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 33 | 311 | 3.6e-93 | 0.9893617021276596 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 1.78e-57 | 35 | 305 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| 225344 | BglC | 2.20e-18 | 34 | 309 | 54 | 364 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.39e-136 | 10 | 335 | 10 | 340 | |
| 2.57e-134 | 10 | 335 | 10 | 340 | |
| 1.22e-132 | 10 | 335 | 10 | 341 | |
| 1.22e-132 | 10 | 335 | 10 | 341 | |
| 2.21e-99 | 31 | 317 | 92 | 397 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.21e-99 | 31 | 319 | 4 | 310 | Chain A, Endoglucanase EG-II [Trichoderma reesei],3QR3_B Chain B, Endoglucanase EG-II [Trichoderma reesei] |
|
| 6.70e-85 | 31 | 319 | 23 | 326 | Structrue of a lucidum protein [Ganoderma lucidum],5D8W_B Structrue of a lucidum protein [Ganoderma lucidum],5D8Z_A Structrue of a lucidum protein [Ganoderma lucidum],5D8Z_B Structrue of a lucidum protein [Ganoderma lucidum] |
|
| 2.83e-51 | 34 | 334 | 7 | 304 | Structure of Thermoascus aurantiacus family 5 endoglucanase [Thermoascus aurantiacus],1GZJ_B Structure of Thermoascus aurantiacus family 5 endoglucanase [Thermoascus aurantiacus] |
|
| 5.67e-51 | 34 | 334 | 8 | 305 | Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus [Thermoascus aurantiacus],1H1N_B Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus [Thermoascus aurantiacus] |
|
| 7.76e-51 | 31 | 317 | 28 | 320 | Crystal structure of the endo-beta-1,4-glucanase Xac0029 from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.17e-133 | 10 | 335 | 10 | 341 | Endoglucanase 1 OS=Saitozyma flava OX=5416 GN=CMC1 PE=2 SV=1 |
|
| 7.31e-98 | 31 | 319 | 94 | 400 | Endoglucanase EG-II OS=Hypocrea jecorina OX=51453 GN=egl2 PE=1 SV=1 |
|
| 2.05e-97 | 31 | 317 | 94 | 398 | Endoglucanase EG-II OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=egl2 PE=1 SV=1 |
|
| 1.30e-92 | 23 | 335 | 69 | 386 | Manganese dependent endoglucanase Eg5A OS=Phanerodontia chrysosporium OX=2822231 GN=Eg5A PE=1 SV=2 |
|
| 7.52e-51 | 7 | 334 | 3 | 328 | Probable endo-beta-1,4-glucanase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
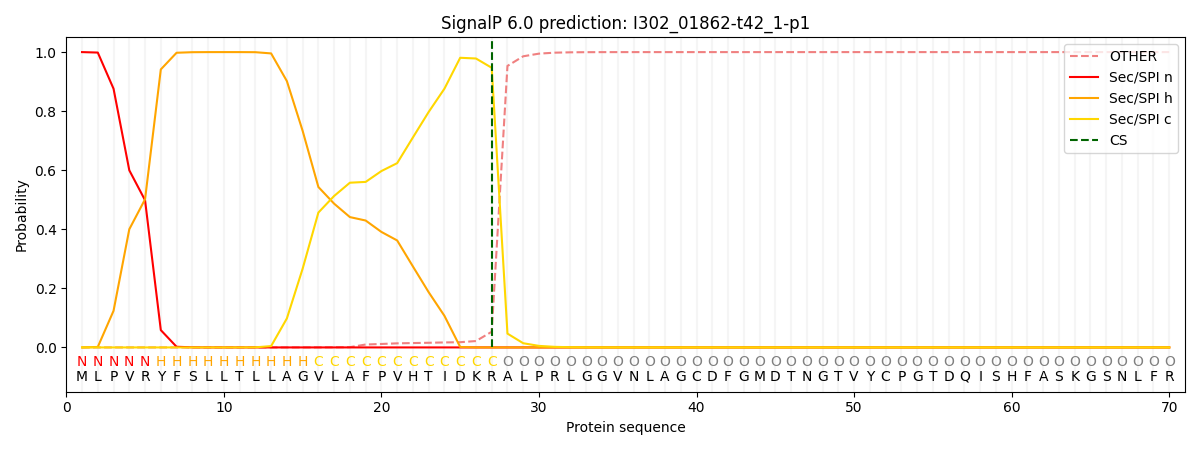
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000321 | 0.999676 | CS pos: 27-28. Pr: 0.9470 |
