You are browsing environment: FUNGIDB
CAZyme Information: HpaG804997:RNA-p1
You are here: Home > Sequence: HpaG804997:RNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
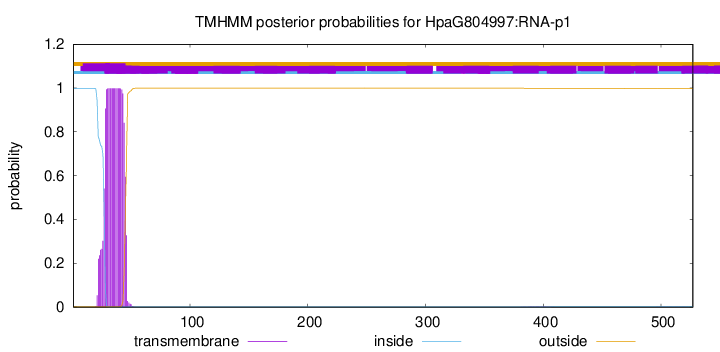
TMHMM annotations
Basic Information help
| Species | Hyaloperonospora arabidopsidis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Hyaloperonospora; Hyaloperonospora arabidopsidis | |||||||||||
| CAZyme ID | HpaG804997:RNA-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 237 | 480 | 1.2e-37 | 0.9701492537313433 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397491 | Anp1 | 1.07e-33 | 237 | 479 | 3 | 262 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 282904 | Herpes_BLLF1 | 3.14e-06 | 59 | 164 | 481 | 587 | Herpes virus major outer envelope glycoprotein (BLLF1). This family consists of the BLLF1 viral late glycoprotein, also termed gp350/220. It is the most abundantly expressed glycoprotein in the viral envelope of the Herpesviruses and is the major antigen responsible for stimulating the production of neutralising antibodies in vivo. |
| 411474 | fibronec_FbpA | 0.001 | 65 | 171 | 181 | 293 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 411408 | PspC_subgroup_2 | 0.005 | 44 | 207 | 133 | 290 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| 411474 | fibronec_FbpA | 0.006 | 59 | 200 | 158 | 277 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.00e-45 | 252 | 525 | 87 | 399 | |
| 3.23e-44 | 256 | 527 | 85 | 380 | |
| 3.23e-44 | 256 | 527 | 85 | 380 | |
| 1.11e-43 | 258 | 524 | 69 | 369 | |
| 2.99e-43 | 242 | 525 | 70 | 385 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.17e-13 | 319 | 525 | 89 | 302 | Crystal structure of Saccharomyces cerevisiae Mnn9 in complex with GDP and Mn. [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.21e-19 | 232 | 525 | 40 | 337 | Mannan polymerase complex subunit mnn9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mnn9 PE=3 SV=1 |
|
| 1.11e-16 | 279 | 525 | 116 | 374 | Mannan polymerase II complex anp1 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=anp1 PE=3 SV=1 |
|
| 5.04e-15 | 325 | 525 | 216 | 421 | Vanadate resistance protein OS=Candida albicans OX=5476 GN=VAN1 PE=3 SV=1 |
|
| 2.17e-14 | 325 | 525 | 309 | 514 | Mannan polymerase I complex VAN1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=VAN1 PE=1 SV=3 |
|
| 2.92e-13 | 319 | 525 | 179 | 392 | Mannan polymerase complexes subunit MNN9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN9 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999987 | 0.000020 |