You are browsing environment: FUNGIDB
CAZyme Information: HVAS_10204500.1-p1
Basic Information
help
| Species |
Hemileia vastatrix
|
| Lineage |
Basidiomycota; Pucciniomycetes; ; Zaghouaniaceae; Hemileia; Hemileia vastatrix
|
| CAZyme ID |
HVAS_10204500.1-p1
|
| CAZy Family |
PL35 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 501 |
|
58022.20 |
8.6524 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_HvastatrixRaceXXXIII |
12854 |
N/A |
284 |
12570
|
|
| Gene Location |
No EC number prediction in HVAS_10204500.1-p1.
HVAS_10204500.1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 2.64e-105 |
16 |
488 |
150 |
633 |
| 4.05e-69 |
97 |
462 |
169 |
556 |
| 1.41e-65 |
49 |
488 |
523 |
974 |
| 1.83e-65 |
97 |
488 |
171 |
573 |
| 1.04e-63 |
97 |
488 |
114 |
522 |
HVAS_10204500.1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.31e-45 |
96 |
476 |
151 |
534 |
Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
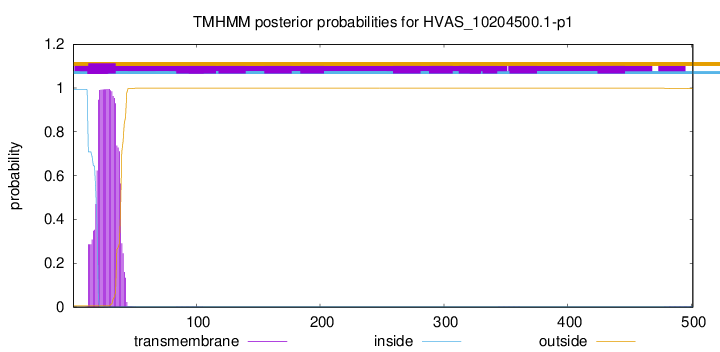
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
CS Position |
| 1.000008 |
0.000003 |
|