You are browsing environment: FUNGIDB
CAZyme Information: HVAS_10157122.1-p1
You are here: Home > Sequence: HVAS_10157122.1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
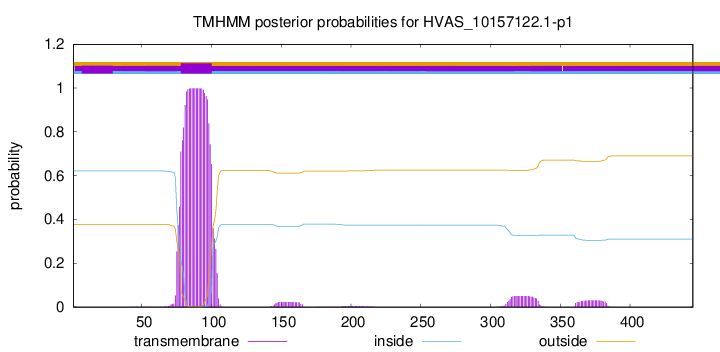
TMHMM annotations
Basic Information help
| Species | Hemileia vastatrix | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Zaghouaniaceae; Hemileia; Hemileia vastatrix | |||||||||||
| CAZyme ID | HVAS_10157122.1-p1 | |||||||||||
| CAZy Family | GH38 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.183:36 | 2.4.1.-:11 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340822 | GT5_Glycogen_synthase_DULL1-like | 1.29e-42 | 174 | 381 | 1 | 208 | Glycogen synthase GlgA and similar proteins. This family is most closely related to the GT5 family of glycosyltransferases. Glycogen synthase (EC:2.4.1.21) catalyzes the formation and elongation of the alpha-1,4-glucose backbone using ADP-glucose, the second and key step of glycogen biosynthesis. This family includes starch synthases of plants, such as DULL1 in Zea mays and glycogen synthases of various organisms. |
| 400563 | Glyco_transf_5 | 3.65e-13 | 175 | 341 | 1 | 175 | Starch synthase catalytic domain. |
| 234809 | glgA | 1.01e-09 | 191 | 341 | 14 | 163 | glycogen synthase GlgA. |
| 223374 | GlgA | 2.17e-07 | 174 | 384 | 2 | 211 | Glycogen synthase [Carbohydrate transport and metabolism]. |
| 172588 | PRK14098 | 3.72e-04 | 304 | 362 | 143 | 211 | starch synthase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.84e-173 | 1 | 445 | 925 | 1364 | |
| 2.41e-168 | 2 | 445 | 1001 | 1439 | |
| 3.58e-168 | 1 | 445 | 811 | 1245 | |
| 8.92e-168 | 2 | 445 | 1001 | 1439 | |
| 1.54e-167 | 2 | 445 | 991 | 1429 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.43e-109 | 21 | 443 | 1011 | 1410 | Cell wall alpha-1,3-glucan synthase ags1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ags1 PE=1 SV=3 |
|
| 9.60e-105 | 4 | 445 | 978 | 1411 | Cell wall alpha-1,3-glucan synthase mok13 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok13 PE=3 SV=2 |
|
| 1.84e-96 | 21 | 424 | 1011 | 1397 | Cell wall alpha-1,3-glucan synthase mok11 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok11 PE=3 SV=2 |
|
| 3.62e-92 | 48 | 436 | 148 | 517 | Cell wall alpha-1,3-glucan synthase mok14 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok14 PE=1 SV=1 |
|
| 1.86e-88 | 1 | 428 | 1006 | 1427 | Cell wall alpha-1,3-glucan synthase mok12 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok12 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000028 | 0.000002 |