You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_12047-t46_1-p1
You are here: Home > Sequence: HMPREF1544_12047-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
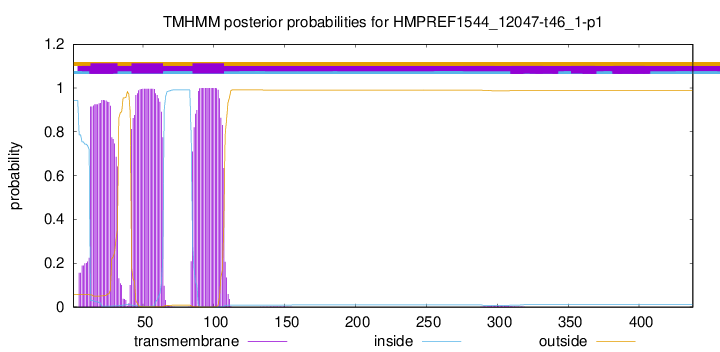
TMHMM annotations
Basic Information help
| Species | Mucor circinelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides | |||||||||||
| CAZyme ID | HMPREF1544_12047-t46_1-p1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.149:9 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 1.81e-21 | 156 | 348 | 1 | 196 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 403517 | EOS1 | 7.00e-07 | 2 | 102 | 25 | 156 | N-glycosylation protein. This family is not required for survival of S.cerevisiae, but its deletion leads to heightened sensitivity to oxidative stress. It appears to be involved in N-glycosylation, and resides in the endoplasmic reticulum. |
| 215596 | PLN03133 | 2.74e-06 | 203 | 337 | 440 | 579 | beta-1,3-galactosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.05e-213 | 1 | 438 | 94 | 522 | |
| 8.06e-127 | 1 | 432 | 91 | 510 | |
| 2.01e-106 | 1 | 435 | 117 | 585 | |
| 1.65e-20 | 137 | 373 | 50 | 286 | |
| 1.93e-19 | 135 | 392 | 13 | 256 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.51e-15 | 136 | 364 | 45 | 275 | Beta-1,3-galactosyltransferase 6 OS=Mus musculus OX=10090 GN=B3galt6 PE=2 SV=1 |
|
| 4.93e-13 | 133 | 396 | 429 | 684 | Hydroxyproline O-galactosyltransferase GALT2 OS=Arabidopsis thaliana OX=3702 GN=GALT2 PE=1 SV=1 |
|
| 1.08e-12 | 133 | 365 | 364 | 596 | Hydroxyproline O-galactosyltransferase GALT3 OS=Arabidopsis thaliana OX=3702 GN=GALT3 PE=2 SV=1 |
|
| 1.76e-12 | 88 | 342 | 7 | 255 | Beta-1,3-galactosyltransferase 5 (Fragment) OS=Gorilla gorilla gorilla OX=9595 GN=B3GALT5 PE=3 SV=2 |
|
| 2.66e-12 | 88 | 342 | 7 | 255 | Beta-1,3-galactosyltransferase 5 OS=Homo sapiens OX=9606 GN=B3GALT5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000019 | 0.000001 |