You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_11450-t46_1-p1
You are here: Home > Sequence: HMPREF1544_11450-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mucor circinelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides | |||||||||||
| CAZyme ID | HMPREF1544_11450-t46_1-p1 | |||||||||||
| CAZy Family | GT57 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.28:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH37 | 52 | 612 | 1.4e-150 | 0.9918533604887984 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395961 | Trehalase | 2.19e-136 | 50 | 605 | 1 | 499 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| 215307 | PLN02567 | 2.99e-123 | 37 | 613 | 13 | 543 | alpha,alpha-trehalase |
| 224541 | TreA | 1.43e-91 | 34 | 607 | 53 | 545 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| 237326 | treA | 2.49e-87 | 18 | 606 | 27 | 528 | alpha,alpha-trehalase TreA. |
| 183936 | treA | 2.75e-83 | 153 | 612 | 142 | 534 | alpha,alpha-trehalase TreA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.30e-227 | 30 | 627 | 28 | 599 | |
| 2.69e-211 | 32 | 624 | 33 | 577 | |
| 1.13e-147 | 32 | 625 | 39 | 647 | |
| 3.04e-147 | 17 | 625 | 15 | 640 | |
| 4.47e-147 | 32 | 625 | 39 | 647 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.71e-76 | 40 | 606 | 15 | 494 | Family 37 trehalase from Escherichia coli in complex with 1- thiatrehazolin [Escherichia coli K-12],2JJB_A Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_B Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_C Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_D Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2WYN_A Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_B Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_C Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_D Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12] |
|
| 1.12e-75 | 34 | 606 | 46 | 531 | Structure of periplasmic trehalase from Diamondback moth gut bacteria complexed with validoxylamine [Enterobacter cloacae],5Z6H_A Structure of periplasmic trehalase from Diamondback moth gut bacteria in the apo form [Enterobacter cloacae],5Z6H_B Structure of periplasmic trehalase from Diamondback moth gut bacteria in the apo form [Enterobacter cloacae] |
|
| 9.20e-72 | 40 | 606 | 15 | 494 | Family 37 trehalase from Escherichia coli in complex with validoxylamine [Escherichia coli K-12] |
|
| 3.08e-70 | 54 | 606 | 40 | 546 | Chain A, Trehalase [Arabidopsis thaliana],7E9U_B Chain B, Trehalase [Arabidopsis thaliana] |
|
| 4.36e-69 | 54 | 606 | 40 | 546 | Chain A, Trehalase [Arabidopsis thaliana],7E9X_B Chain B, Trehalase [Arabidopsis thaliana],7E9X_C Chain C, Trehalase [Arabidopsis thaliana],7E9X_D Chain D, Trehalase [Arabidopsis thaliana],7EAW_A Chain A, Trehalase [Arabidopsis thaliana],7EAW_B Chain B, Trehalase [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.36e-133 | 32 | 622 | 49 | 594 | Trehalase OS=Dictyostelium discoideum OX=44689 GN=treh PE=3 SV=1 |
|
| 1.26e-107 | 32 | 621 | 45 | 578 | Trehalase OS=Pimpla hypochondriaca OX=135724 GN=tre1 PE=1 SV=1 |
|
| 2.97e-106 | 32 | 616 | 25 | 549 | Trehalase OS=Mus musculus OX=10090 GN=Treh PE=1 SV=1 |
|
| 1.52e-103 | 31 | 613 | 24 | 551 | Trehalase OS=Tenebrio molitor OX=7067 PE=2 SV=1 |
|
| 2.89e-103 | 32 | 629 | 28 | 563 | Trehalase OS=Oryctolagus cuniculus OX=9986 GN=TREH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
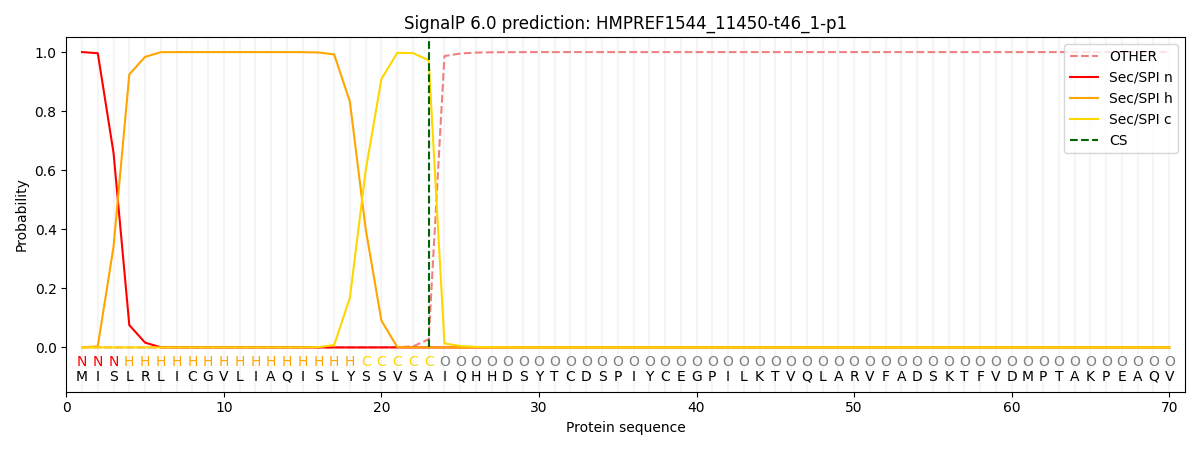
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000210 | 0.999799 | CS pos: 23-24. Pr: 0.9725 |
