You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_08552-t46_1-p1
You are here: Home > Sequence: HMPREF1544_08552-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
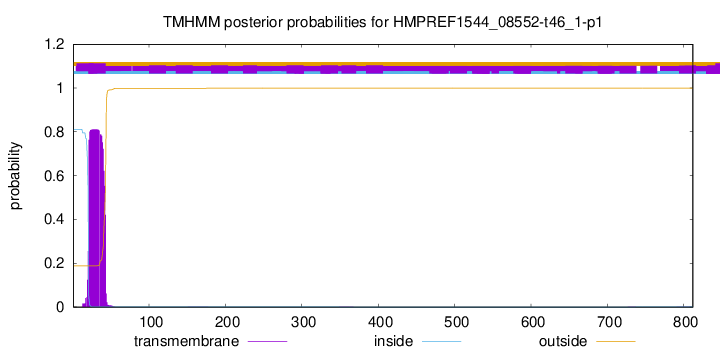
TMHMM annotations
Basic Information help
| Species | Mucor circinelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides | |||||||||||
| CAZyme ID | HMPREF1544_08552-t46_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.106:25 | 3.2.1.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH63 | 596 | 810 | 7.3e-28 | 0.43333333333333335 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397353 | Glyco_hydro_63 | 0.0 | 311 | 810 | 1 | 494 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| 407154 | Glyco_hydro_63N | 8.77e-81 | 49 | 245 | 1 | 200 | Glycosyl hydrolase family 63 N-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the N-terminal beta sandwich domain. |
| 225942 | GDB1 | 4.02e-09 | 614 | 762 | 435 | 565 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| 283786 | GDE_C | 4.44e-05 | 625 | 729 | 191 | 300 | Amylo-alpha-1,6-glucosidase. This family includes human glycogen branching enzyme AGL. This enzyme contains a number of distinct catalytic activities. It has been shown for the yeast homolog GDB1 that mutations in this region disrupt the enzymes Amylo-alpha-1,6-glucosidase (EC:3.2.1.33). |
| 395961 | Trehalase | 0.001 | 599 | 791 | 306 | 483 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 44 | 812 | 37 | 804 | |
| 1.43e-248 | 42 | 812 | 18 | 771 | |
| 6.36e-247 | 46 | 812 | 35 | 801 | |
| 1.09e-241 | 13 | 812 | 2 | 807 | |
| 1.44e-241 | 75 | 812 | 1 | 744 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.00e-159 | 45 | 810 | 11 | 802 | Crystal structure of Processing alpha-Glucosidase I [Saccharomyces cerevisiae S288C] |
|
| 6.76e-149 | 49 | 812 | 37 | 779 | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_B Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_C Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_D Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.20e-219 | 44 | 812 | 37 | 808 | Probable mannosyl-oligosaccharide glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC6G10.09 PE=3 SV=1 |
|
| 1.46e-158 | 10 | 810 | 3 | 832 | Mannosyl-oligosaccharide glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CWH41 PE=1 SV=1 |
|
| 9.34e-153 | 49 | 812 | 92 | 834 | Mannosyl-oligosaccharide glucosidase OS=Mus musculus OX=10090 GN=Mogs PE=1 SV=1 |
|
| 2.21e-147 | 49 | 812 | 94 | 837 | Mannosyl-oligosaccharide glucosidase OS=Homo sapiens OX=9606 GN=MOGS PE=1 SV=5 |
|
| 1.02e-143 | 51 | 812 | 94 | 834 | Mannosyl-oligosaccharide glucosidase OS=Rattus norvegicus OX=10116 GN=Mogs PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000886 | 0.999070 | CS pos: 35-36. Pr: 0.9728 |