You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_07070-t46_1-p1
You are here: Home > Sequence: HMPREF1544_07070-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
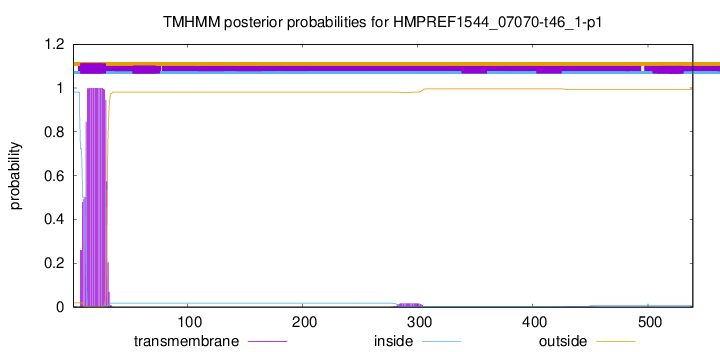
TMHMM annotations
Basic Information help
| Species | Mucor circinelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides | |||||||||||
| CAZyme ID | HMPREF1544_07070-t46_1-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 212 | 453 | 2.8e-52 | 0.9772727272727273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 402574 | Mannosyl_trans3 | 2.11e-48 | 213 | 456 | 3 | 273 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
| 129661 | cdk7 | 0.009 | 31 | 96 | 131 | 193 | CDK-activating kinase assembly factor MAT1. All proteins in this family for which functions are known are cyclin dependent protein kinases that are components of TFIIH, a complex that is involved in nucleotide excision repair and transcription initiation. Also known as MAT1 (menage a trois 1). This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.55e-206 | 6 | 527 | 6 | 517 | |
| 2.10e-193 | 6 | 526 | 3 | 514 | |
| 7.70e-56 | 135 | 520 | 92 | 499 | |
| 1.37e-53 | 136 | 512 | 102 | 499 | |
| 1.87e-53 | 163 | 476 | 197 | 554 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.40e-18 | 199 | 439 | 262 | 521 | Putative alpha-1,3-mannosyltransferase MNN14 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN14 PE=2 SV=1 |
|
| 4.79e-16 | 141 | 512 | 254 | 660 | Alpha-1,3-mannosyltransferase MNN1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN1 PE=1 SV=1 |
|
| 2.66e-15 | 133 | 439 | 275 | 624 | Putative alpha-1,3-mannosyltransferase MNN1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN1 PE=2 SV=1 |
|
| 1.37e-14 | 197 | 479 | 161 | 474 | Alpha-1,3-mannosyltransferase MNT2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT2 PE=1 SV=1 |
|
| 3.64e-14 | 233 | 468 | 255 | 526 | Alpha-1,3-mannosyltransferase MNT3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999967 | 0.000070 |