You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_03901-t46_1-p1
You are here: Home > Sequence: HMPREF1544_03901-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
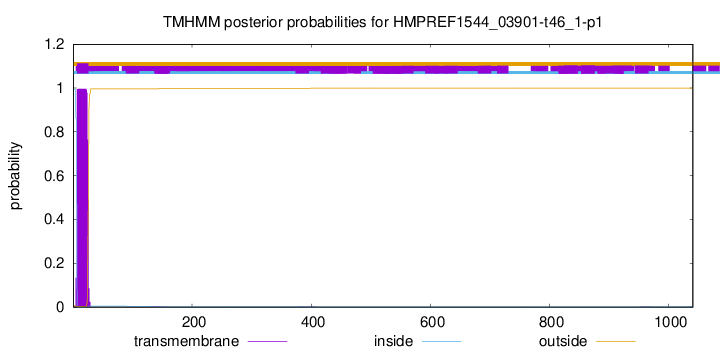
TMHMM annotations
Basic Information help
| Species | Mucor circinelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides | |||||||||||
| CAZyme ID | HMPREF1544_03901-t46_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 58 | 438 | 7.2e-76 | 0.9869706840390879 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 9.57e-55 | 57 | 426 | 1 | 302 | Glycosyl hydrolases family 35. |
| 166698 | PLN03059 | 1.23e-18 | 53 | 450 | 32 | 357 | beta-galactosidase; Provisional |
| 224786 | GanA | 2.92e-18 | 54 | 219 | 4 | 160 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 7.37e-06 | 79 | 220 | 8 | 138 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 5 | 1041 | 3 | 1033 | |
| 1.12e-76 | 51 | 1018 | 39 | 933 | |
| 6.54e-73 | 50 | 1006 | 43 | 942 | |
| 2.18e-72 | 50 | 1006 | 43 | 942 | |
| 1.06e-71 | 36 | 1018 | 27 | 936 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.71e-43 | 51 | 1020 | 6 | 956 | Native structure of beta-galactosidase from Penicillium sp. [Penicillium sp.],1XC6_A Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose [Penicillium sp.] |
|
| 3.06e-43 | 51 | 988 | 46 | 955 | Structure Of Beta-galactosidase From Aspergillus Niger [Aspergillus niger CBS 513.88] |
|
| 7.06e-43 | 51 | 988 | 46 | 955 | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 3-b-Galactopyranosyl glucose [Aspergillus niger CBS 513.88],5IHR_A Structure Of E298q-beta-galactosidase From Aspergillus Niger In Complex With Allolactose [Aspergillus niger CBS 513.88],5JUV_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-b-Galactopyranosyl galactose [Aspergillus niger CBS 513.88],5MGC_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 4-Galactosyl-lactose [Aspergillus niger CBS 513.88],5MGD_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose [Aspergillus niger CBS 513.88] |
|
| 2.60e-41 | 44 | 1029 | 39 | 996 | Crystal structure of beta-galactosidase from Aspergillus oryzae in complex with galactose [Aspergillus oryzae] |
|
| 4.51e-41 | 51 | 424 | 26 | 350 | Chain A, Beta-galactosidase [Trichoderma reesei],3OGR_A Chain A, Beta-galactosidase [Trichoderma reesei],3OGS_A Chain A, Beta-galactosidase [Trichoderma reesei],3OGV_A Chain A, Beta-galactosidase [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.04e-52 | 50 | 529 | 36 | 476 | Probable beta-galactosidase B OS=Sclerotinia sclerotiorum (strain ATCC 18683 / 1980 / Ss-1) OX=665079 GN=lacB PE=3 SV=1 |
|
| 1.23e-52 | 51 | 1006 | 32 | 948 | Probable beta-galactosidase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=lacC PE=3 SV=1 |
|
| 2.90e-52 | 51 | 1006 | 32 | 948 | Probable beta-galactosidase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=lacC PE=3 SV=1 |
|
| 2.90e-52 | 51 | 1006 | 32 | 948 | Probable beta-galactosidase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=lacC PE=3 SV=1 |
|
| 7.67e-52 | 50 | 529 | 36 | 476 | Probable beta-galactosidase B OS=Botryotinia fuckeliana (strain B05.10) OX=332648 GN=lacB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
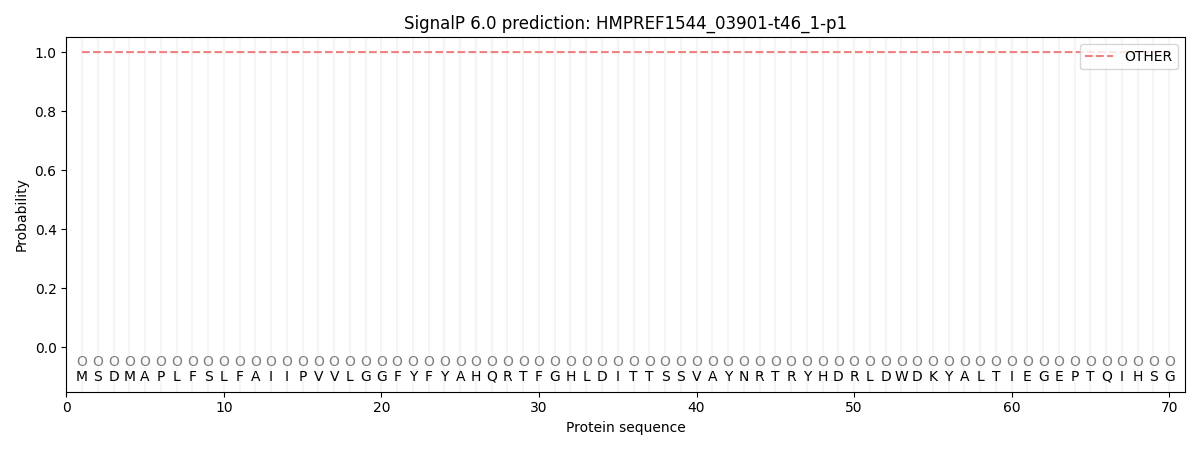
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999978 | 0.000071 |