You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_03268-t46_1-p1
You are here: Home > Sequence: HMPREF1544_03268-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
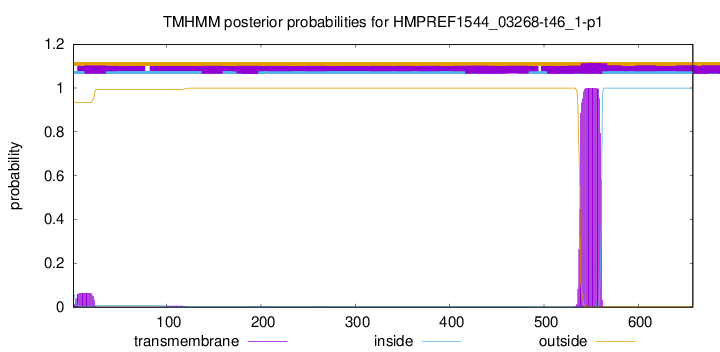
TMHMM annotations
Basic Information help
| Species | Mucor circinelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides | |||||||||||
| CAZyme ID | HMPREF1544_03268-t46_1-p1 | |||||||||||
| CAZy Family | GH152 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 41 | 502 | 1.2e-106 | 0.9976076555023924 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395616 | Glyco_hydro_9 | 8.41e-122 | 44 | 501 | 1 | 374 | Glycosyl hydrolase family 9. |
| 177732 | PLN00119 | 3.20e-87 | 29 | 502 | 19 | 486 | endoglucanase |
| 177979 | PLN02345 | 1.73e-85 | 46 | 494 | 2 | 447 | endoglucanase |
| 215331 | PLN02613 | 1.42e-82 | 39 | 497 | 24 | 471 | endoglucanase |
| 215194 | PLN02340 | 1.16e-81 | 31 | 496 | 20 | 485 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.66e-123 | 35 | 540 | 17 | 496 | |
| 1.07e-121 | 17 | 552 | 6 | 512 | |
| 6.12e-120 | 27 | 514 | 32 | 512 | |
| 1.71e-119 | 27 | 514 | 32 | 512 | |
| 7.70e-117 | 52 | 565 | 3 | 482 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.42e-88 | 38 | 505 | 2 | 441 | EndoEXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA [Thermobifida fusca],1JS4_B EndoEXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA [Thermobifida fusca],1TF4_A EndoEXOCELLULASE FROM THERMOMONOSPORA [Thermobifida fusca],1TF4_B EndoEXOCELLULASE FROM THERMOMONOSPORA [Thermobifida fusca],3TF4_A EndoEXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA [Thermobifida fusca],3TF4_B EndoEXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA [Thermobifida fusca],4TF4_A EndoEXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA [Thermobifida fusca],4TF4_B EndoEXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA [Thermobifida fusca] |
|
| 1.63e-80 | 41 | 502 | 4 | 426 | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 2.5. [Nasutitermes takasagoensis],1KSC_A The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 5.6. [Nasutitermes takasagoensis],1KSD_A The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 6.5. [Nasutitermes takasagoensis] |
|
| 7.96e-68 | 42 | 502 | 8 | 428 | Chain A, Endo-1, 4-beta-glucanase [Eisenia fetida] |
|
| 1.36e-66 | 42 | 512 | 6 | 444 | The X-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum],1K72_B The X-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum] |
|
| 1.89e-66 | 42 | 512 | 6 | 444 | The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1G87_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1GA2_A The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1GA2_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1KFG_A The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum],1KFG_B The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.39e-85 | 38 | 505 | 48 | 487 | Endoglucanase E-4 OS=Thermobifida fusca OX=2021 GN=celD PE=1 SV=2 |
|
| 9.74e-85 | 39 | 510 | 46 | 488 | Endoglucanase A OS=Bacillus pumilus OX=1408 GN=eglA PE=1 SV=1 |
|
| 5.47e-80 | 29 | 502 | 21 | 488 | Endoglucanase 14 OS=Arabidopsis thaliana OX=3702 GN=At2g44560 PE=2 SV=2 |
|
| 1.86e-79 | 42 | 497 | 30 | 456 | Endoglucanase 4 OS=Bacillus sp. (strain KSM-522) OX=120046 PE=3 SV=2 |
|
| 1.62e-77 | 31 | 502 | 23 | 487 | Endoglucanase 13 OS=Arabidopsis thaliana OX=3702 GN=At2g44550 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
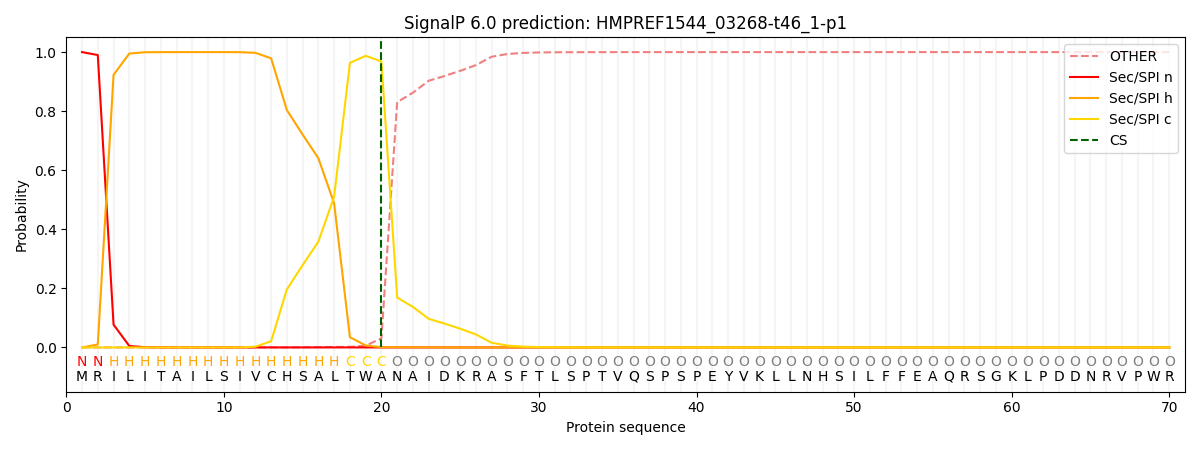
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000307 | 0.999670 | CS pos: 20-21. Pr: 0.9684 |