You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_01234-t46_1-p1
Basic Information
help
| Species |
Mucor circinelloides
|
| Lineage |
Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides
|
| CAZyme ID |
HMPREF1544_01234-t46_1-p1
|
| CAZy Family |
CBM21 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 545 |
|
60340.97 |
4.2793 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_Mcircinelloides1006PhL |
12410 |
1220926 |
183 |
12227
|
|
| Gene Location |
No EC number prediction in HMPREF1544_01234-t46_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA12 |
91 |
489 |
7.4e-36 |
0.9775561097256857 |
HMPREF1544_01234-t46_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.92e-36 |
97 |
495 |
60 |
466 |
| 4.20e-22 |
162 |
490 |
163 |
507 |
| 4.23e-22 |
162 |
490 |
164 |
508 |
| 2.15e-21 |
97 |
490 |
38 |
452 |
| 7.20e-21 |
89 |
492 |
28 |
422 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.31e-08 |
99 |
492 |
16 |
403 |
Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
| 1.31e-08 |
99 |
492 |
15 |
402 |
Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
| 1.32e-08 |
99 |
492 |
17 |
404 |
Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
HMPREF1544_01234-t46_1-p1 has no Swissprot hit.
This protein is predicted as SP
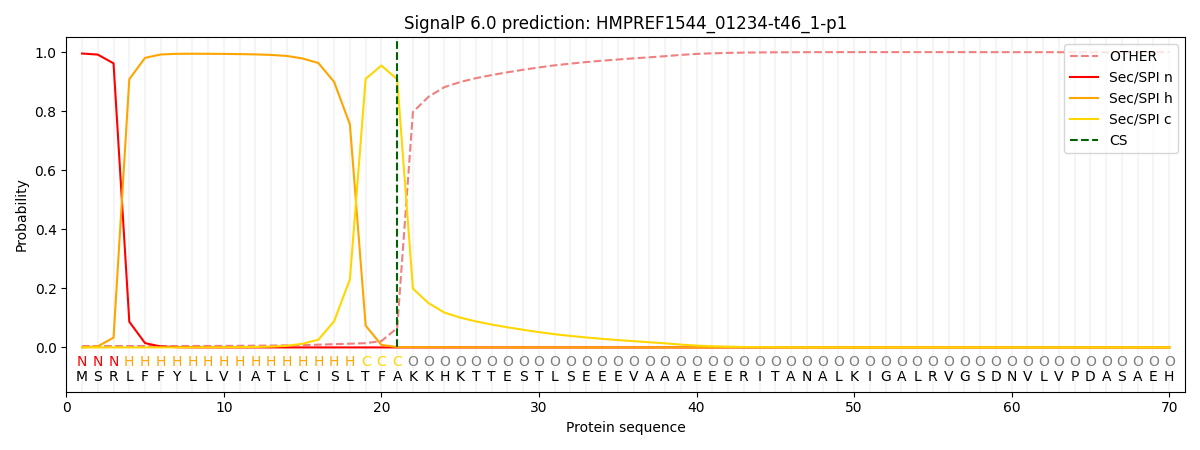
| Other |
SP_Sec_SPI |
CS Position |
| 0.006704 |
0.993262 |
CS pos: 21-22. Pr: 0.9083 |
There is no transmembrane helices in HMPREF1544_01234-t46_1-p1.

