You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_00296-t46_1-p1
Basic Information
help
| Species |
Mucor circinelloides
|
| Lineage |
Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides
|
| CAZyme ID |
HMPREF1544_00296-t46_1-p1
|
| CAZy Family |
AA2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 362 |
KE123897|CGC1 |
41542.76 |
6.7677 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_Mcircinelloides1006PhL |
12410 |
1220926 |
183 |
12227
|
|
| Gene Location |
No EC number prediction in HMPREF1544_00296-t46_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT77 |
145 |
354 |
4e-21 |
0.9814814814814815 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 367480
|
Nucleotid_trans |
1.34e-14 |
141 |
354 |
1 |
208 |
Nucleotide-diphospho-sugar transferase. Proteins in this family have been been predicted to be nucleotide-diphospho-sugar transferases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 8.03e-139 |
1 |
362 |
1 |
356 |
| 1.14e-136 |
1 |
362 |
1 |
368 |
| 8.63e-76 |
41 |
362 |
33 |
333 |
| 8.38e-71 |
115 |
362 |
127 |
370 |
| 2.54e-65 |
110 |
362 |
109 |
353 |
HMPREF1544_00296-t46_1-p1 has no PDB hit.
HMPREF1544_00296-t46_1-p1 has no Swissprot hit.
This protein is predicted as SP
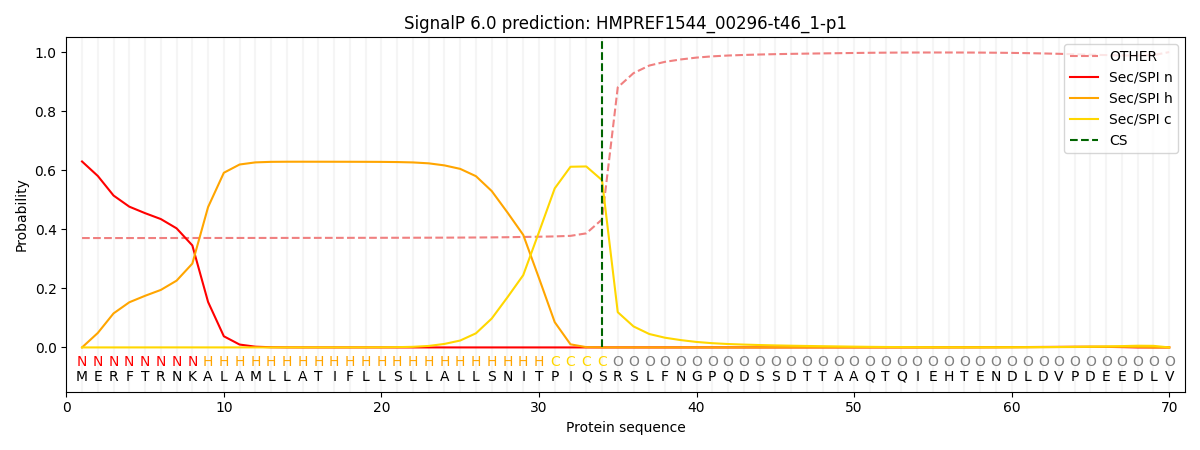
| Other |
SP_Sec_SPI |
CS Position |
| 0.402584 |
0.597392 |
CS pos: 34-35. Pr: 0.5665 |


