You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1544_00140-t46_1-p1
You are here: Home > Sequence: HMPREF1544_00140-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mucor circinelloides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor circinelloides | |||||||||||
| CAZyme ID | HMPREF1544_00140-t46_1-p1 | |||||||||||
| CAZy Family | AA12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 18 | 420 | 6.4e-102 | 0.9974293059125964 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215437 | PLN02816 | 3.32e-112 | 18 | 511 | 41 | 541 | mannosyltransferase |
| 281842 | Glyco_transf_22 | 1.25e-83 | 18 | 420 | 2 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.35e-149 | 74 | 512 | 1 | 434 | |
| 4.44e-114 | 3 | 514 | 29 | 556 | |
| 4.44e-114 | 3 | 514 | 29 | 556 | |
| 3.59e-113 | 18 | 511 | 41 | 541 | |
| 3.59e-113 | 18 | 511 | 41 | 541 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.38e-114 | 18 | 511 | 41 | 541 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
|
| 1.12e-107 | 18 | 511 | 18 | 508 | GPI mannosyltransferase 3 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=GPI10 PE=3 SV=1 |
|
| 1.95e-104 | 26 | 502 | 71 | 532 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
|
| 1.96e-104 | 16 | 502 | 50 | 521 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
|
| 3.78e-104 | 26 | 502 | 61 | 522 | GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000035 | 0.000006 |
TMHMM Annotations download full data without filtering help
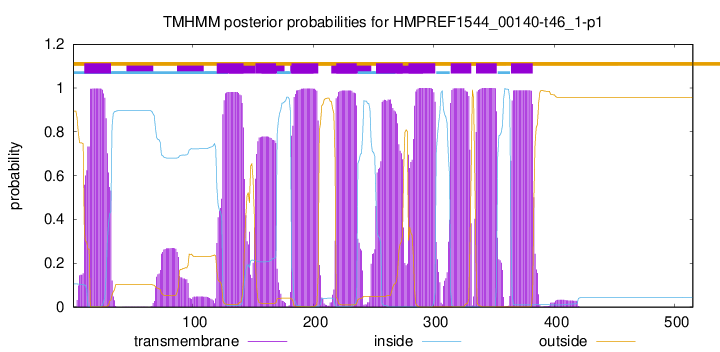
| Start | End |
|---|---|
| 10 | 32 |
| 120 | 142 |
| 152 | 169 |
| 182 | 204 |
| 219 | 236 |
| 252 | 274 |
| 279 | 301 |
| 314 | 331 |
| 335 | 352 |
| 364 | 382 |
