You are browsing environment: FUNGIDB
HMPREF1541_10415-t46_1-p1
Basic Information
help
Species
Cyphellophora europaea
Lineage
Ascomycota; Eurotiomycetes; ; Cyphellophoraceae; Cyphellophora; Cyphellophora europaea
CAZyme ID
HMPREF1541_10415-t46_1-p1
CAZy Family
GT45
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
777
KB822714|CGC2 89397.13
4.5305
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CeuropaeaCBS101466
11153
1220924
59
11094
Gene Location
No EC number prediction in HMPREF1541_10415-t46_1-p1.
Family
Start
End
Evalue
family coverage
GH139
5
775
1.6e-276
0.9806949806949807
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
408731
DUF5703_N
7.19e-144
28
311
1
287
Domain of unknown function (DUF5703). This is an N-terminal domain of unknown function mostly found in bacteria. It is possible that this domain might be a putative glycoside hydrolase. This family belongs to the Galactose Mutarotase-like superfamily.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
2.12e-294
16
777
17
777
3.84e-292
21
777
22
776
7.99e-292
16
777
17
777
7.99e-292
16
777
17
777
4.71e-291
21
777
4
758
HMPREF1541_10415-t46_1-p1 has no PDB hit.
HMPREF1541_10415-t46_1-p1 has no Swissprot hit.
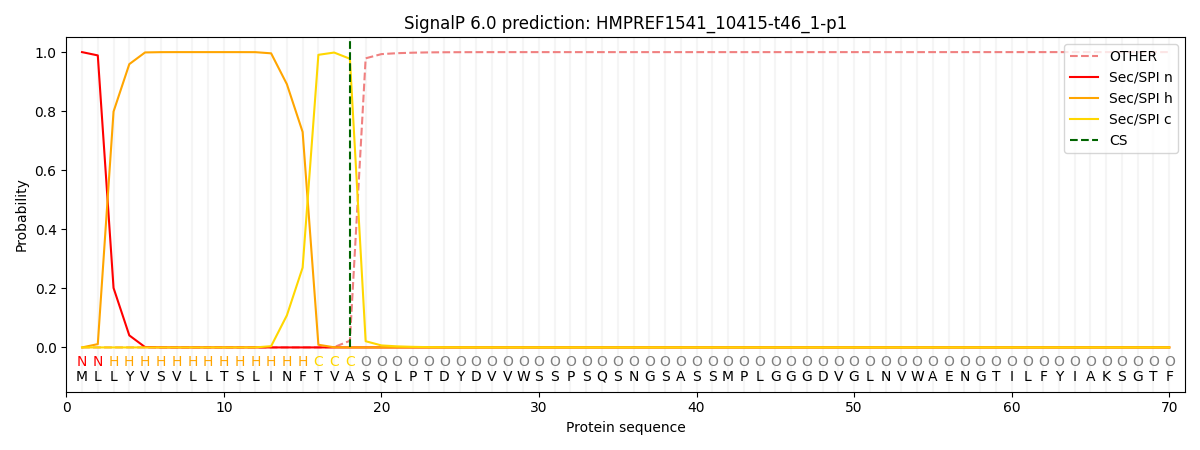
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000208
0.999743
CS pos: 18-19. Pr: 0.9775
There is no transmembrane helices in HMPREF1541_10415-t46_1-p1.