You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1541_09928-t46_1-p1
You are here: Home > Sequence: HMPREF1541_09928-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cyphellophora europaea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Cyphellophoraceae; Cyphellophora; Cyphellophora europaea | |||||||||||
| CAZyme ID | HMPREF1541_09928-t46_1-p1 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.183:18 | 2.4.1.-:2 | 2.4.1.183:36 | 2.4.1.-:11 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH13 | 95 | 491 | 1.4e-154 | 0.9975 |
| GH13 | 1157 | 1621 | 1.4e-64 | 0.98 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 200462 | AmyAc_AGS | 0.0 | 4 | 570 | 2 | 569 | Alpha amylase catalytic domain found in Alpha 1,3-glucan synthase (also called uridine diphosphoglucose-1,3-alpha-glucan glucosyltransferase and 1,3-alpha-D-glucan synthase). Alpha 1,3-glucan synthase (AGS, EC 2.4.1.183) is an enzyme that catalyzes the reversible chemical reaction of UDP-glucose and [alpha-D-glucosyl-(1-3)]n to form UDP and [alpha-D-glucosyl-(1-3)]n+1. AGS is a component of fungal cell walls. The cell wall of filamentous fungi is composed of 10-15% chitin and 10-35% alpha-1,3-glucan. AGS is triggered in fungi as a response to cell wall stress and elongates the glucan chains in cell wall synthesis. This group includes proteins from Ascomycetes and Basidomycetes. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| 340822 | GT5_Glycogen_synthase_DULL1-like | 5.93e-91 | 1158 | 1614 | 2 | 459 | Glycogen synthase GlgA and similar proteins. This family is most closely related to the GT5 family of glycosyltransferases. Glycogen synthase (EC:2.4.1.21) catalyzes the formation and elongation of the alpha-1,4-glucose backbone using ADP-glucose, the second and key step of glycogen biosynthesis. This family includes starch synthases of plants, such as DULL1 in Zea mays and glycogen synthases of various organisms. |
| 200489 | AmyAc_5 | 2.83e-24 | 63 | 490 | 5 | 395 | Alpha amylase catalytic domain found in an uncharacterized protein family. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| 223443 | AmyA | 2.15e-21 | 66 | 596 | 9 | 474 | Glycosidase [Carbohydrate transport and metabolism]. |
| 223374 | GlgA | 4.26e-19 | 1238 | 1564 | 82 | 402 | Glycogen synthase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 8 | 2330 | 7 | 2244 | |
| 0.0 | 17 | 2329 | 26 | 2378 | |
| 0.0 | 17 | 2328 | 21 | 2448 | |
| 0.0 | 19 | 2328 | 23 | 2405 | |
| 0.0 | 19 | 2328 | 23 | 2405 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.14e-14 | 1287 | 1567 | 130 | 403 | Chain A, Glycogen synthase [Escherichia coli] |
|
| 7.39e-14 | 1287 | 1567 | 130 | 403 | Crystal Structure of Wild-type E.coli GS in complex with ADP and Glucose(wtGSb) [Escherichia coli],2R4T_A Crystal Structure of Wild-type E.coli GS in Complex with ADP and Glucose(wtGSc) [Escherichia coli],2R4U_A Crystal Structure of Wild-type E.coli GS in complex with ADP and Glucose(wtGSd) [Escherichia coli],3GUH_A Crystal Structure of Wild-type E.coli GS in complex with ADP and DGM [Escherichia coli K-12] |
|
| 3.95e-13 | 1287 | 1567 | 130 | 403 | Chain A, Glycogen synthase [Escherichia coli],3CX4_A Chain A, Glycogen synthase [Escherichia coli] |
|
| 5.61e-12 | 1166 | 1583 | 18 | 449 | Crystal Structure of Rice Granule bound Starch Synthase I Catalytic Domain [Oryza sativa Japonica Group],3VUF_A Crystal Structure of Rice Granule bound Starch Synthase I Catalytic Domain in Complex with ADP [Oryza sativa Japonica Group] |
|
| 7.68e-11 | 1444 | 1618 | 361 | 547 | Granule Bound Starch Synthase I from Cyanophora paradoxa bound to acarbose and ADP [Cyanophora paradoxa],6GNG_B Granule Bound Starch Synthase I from Cyanophora paradoxa bound to acarbose and ADP [Cyanophora paradoxa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 18 | 2329 | 23 | 2347 | Cell wall alpha-1,3-glucan synthase mok13 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok13 PE=3 SV=2 |
|
| 0.0 | 6 | 2329 | 16 | 2399 | Cell wall alpha-1,3-glucan synthase ags1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ags1 PE=1 SV=3 |
|
| 0.0 | 4 | 2329 | 10 | 2342 | Cell wall alpha-1,3-glucan synthase mok12 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok12 PE=3 SV=1 |
|
| 0.0 | 2 | 2328 | 3 | 2385 | Cell wall alpha-1,3-glucan synthase mok11 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok11 PE=3 SV=2 |
|
| 1.39e-252 | 1077 | 2334 | 186 | 1361 | Cell wall alpha-1,3-glucan synthase mok14 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok14 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
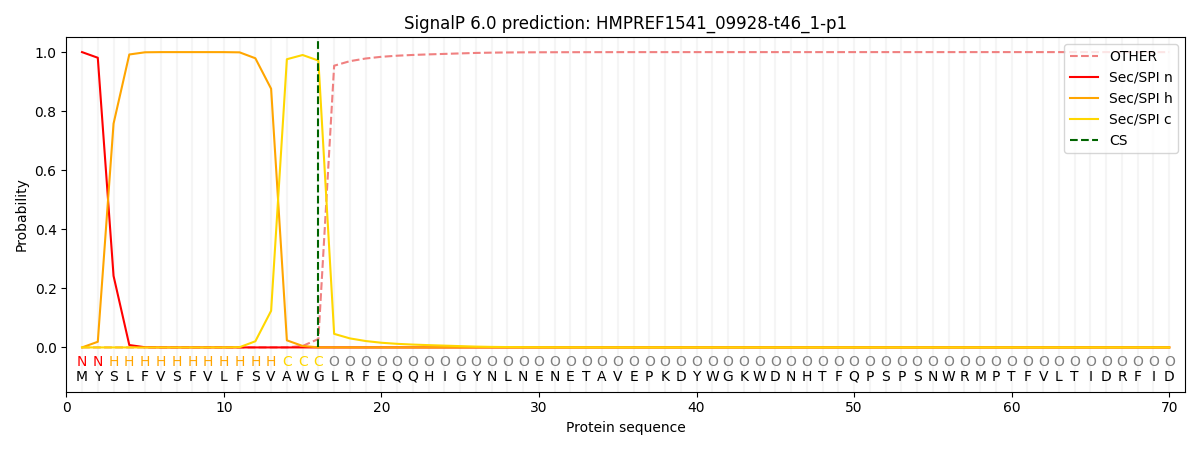
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000750 | 0.999224 | CS pos: 16-17. Pr: 0.9713 |
TMHMM Annotations download full data without filtering help

| Start | End |
|---|---|
| 2310 | 2329 |
| 1947 | 1969 |
| 1976 | 1998 |
| 2008 | 2030 |
| 2043 | 2065 |
| 2085 | 2107 |
| 2127 | 2146 |
| 2166 | 2188 |
| 2195 | 2217 |
| 2241 | 2263 |
| 2268 | 2290 |
