You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1541_04799-t46_1-p1
You are here: Home > Sequence: HMPREF1541_04799-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
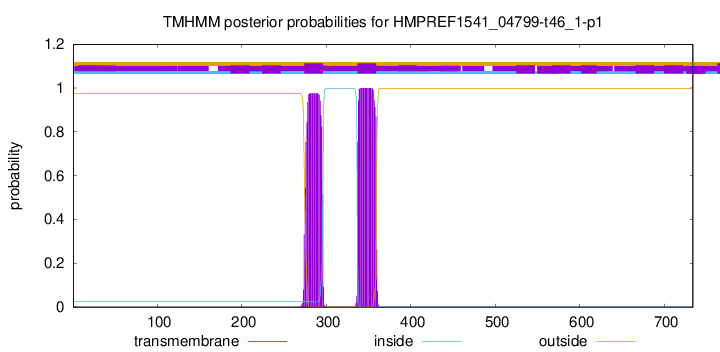
TMHMM annotations
Basic Information help
| Species | Cyphellophora europaea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Cyphellophoraceae; Cyphellophora; Cyphellophora europaea | |||||||||||
| CAZyme ID | HMPREF1541_04799-t46_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 435 | 710 | 1.3e-33 | 0.9517684887459807 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 2.89e-37 | 438 | 687 | 60 | 288 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 1.75e-11 | 442 | 710 | 25 | 304 | Glycosyl hydrolases family 17. |
| 185594 | PTZ00395 | 2.15e-05 | 144 | 243 | 417 | 510 | Sec24-related protein; Provisional |
| 252669 | Extensin_2 | 0.003 | 15 | 87 | 1 | 54 | Extensin-like region. |
| 223039 | PHA03307 | 0.005 | 16 | 219 | 103 | 344 | transcriptional regulator ICP4; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.11e-195 | 75 | 722 | 2 | 614 | |
| 1.21e-190 | 95 | 722 | 79 | 683 | |
| 1.21e-190 | 95 | 722 | 79 | 683 | |
| 1.21e-190 | 95 | 722 | 79 | 683 | |
| 6.19e-189 | 95 | 722 | 10 | 613 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.01e-29 | 426 | 715 | 38 | 294 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
|
| 4.36e-28 | 426 | 715 | 38 | 294 | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose [Rhizomucor miehei CAU432],4WTS_A Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.15e-191 | 95 | 722 | 79 | 683 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgC PE=3 SV=2 |
|
| 2.15e-191 | 95 | 722 | 79 | 683 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgC PE=3 SV=1 |
|
| 9.73e-187 | 83 | 722 | 51 | 693 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgC PE=3 SV=1 |
|
| 2.19e-186 | 102 | 722 | 80 | 686 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgC PE=3 SV=1 |
|
| 4.39e-186 | 102 | 722 | 80 | 686 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000051 | 0.000000 |