You are browsing environment: FUNGIDB
CAZyme Information: HMPREF1541_04676-t46_1-p1
You are here: Home > Sequence: HMPREF1541_04676-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cyphellophora europaea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Cyphellophoraceae; Cyphellophora; Cyphellophora europaea | |||||||||||
| CAZyme ID | HMPREF1541_04676-t46_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.106:25 | 3.2.1.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH63 | 572 | 785 | 1.9e-30 | 0.4298245614035088 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397353 | Glyco_hydro_63 | 0.0 | 303 | 787 | 1 | 494 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| 407154 | Glyco_hydro_63N | 6.38e-103 | 44 | 261 | 1 | 221 | Glycosyl hydrolase family 63 N-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the N-terminal beta sandwich domain. |
| 225942 | GDB1 | 8.92e-07 | 290 | 716 | 159 | 549 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| 236653 | PRK10137 | 0.001 | 658 | 779 | 667 | 773 | alpha-glucosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 39 | 804 | 39 | 822 | |
| 0.0 | 39 | 804 | 39 | 822 | |
| 0.0 | 39 | 794 | 43 | 810 | |
| 0.0 | 39 | 804 | 39 | 822 | |
| 0.0 | 39 | 804 | 39 | 822 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.04e-164 | 41 | 785 | 12 | 800 | Crystal structure of Processing alpha-Glucosidase I [Saccharomyces cerevisiae S288C] |
|
| 9.09e-125 | 44 | 788 | 37 | 778 | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_B Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_C Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_D Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.86e-232 | 12 | 786 | 19 | 805 | Probable mannosyl-oligosaccharide glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC6G10.09 PE=3 SV=1 |
|
| 1.98e-163 | 41 | 785 | 42 | 830 | Mannosyl-oligosaccharide glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CWH41 PE=1 SV=1 |
|
| 2.32e-124 | 44 | 788 | 92 | 833 | Mannosyl-oligosaccharide glucosidase OS=Mus musculus OX=10090 GN=Mogs PE=1 SV=1 |
|
| 1.01e-121 | 44 | 788 | 92 | 833 | Mannosyl-oligosaccharide glucosidase OS=Rattus norvegicus OX=10116 GN=Mogs PE=1 SV=1 |
|
| 1.65e-120 | 40 | 786 | 105 | 846 | Mannosyl-oligosaccharide glucosidase GCS1 OS=Arabidopsis thaliana OX=3702 GN=GCS1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
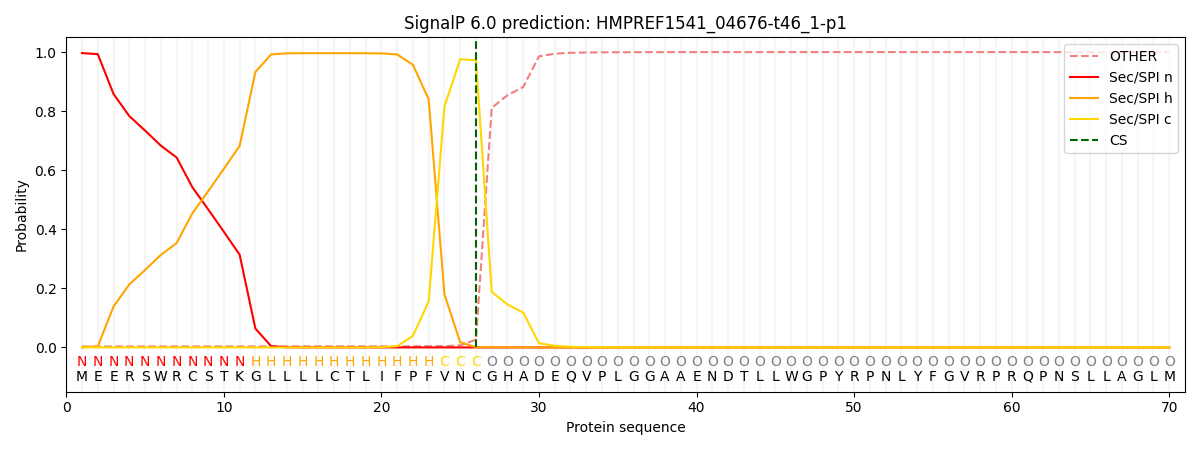
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.006642 | 0.993327 | CS pos: 26-27. Pr: 0.9726 |
