You are browsing environment: FUNGIDB
CAZyme Information: HCEG_02405-t36_1-p1
You are here: Home > Sequence: HCEG_02405-t36_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | HCEG_02405-t36_1-p1 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | proteasome activator subunit 4 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT76 | 30 | 448 | 6.5e-93 | 0.914004914004914 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 406818 | BLM10_mid | 0.0 | 900 | 1427 | 1 | 499 | Proteasome-substrate-size regulator, mid region. The ordered regions of the yeast BLM10 or PA200 (human homolog), full-length protein encode 32 HEAT repeat (HR)-like modules, each comprising two helices joined by a turn, with adjacent repeats connected by a linker. Whereas a standard HEAT repeat is composed of ~50 residues, the BLM10 HEAT repeats are highly variable. The length of helices ranges from 8 to 35 residues, turns range from 2 to 87 residues, and linkers range from 1 to 88 residues, with the longest linker, between HR21 and HR22, containing additional secondary structures (two strands and three helices). BLM10_mid is the middle ordered region of the three in BLM10. BLM10 is found to surround the proteasome entry pore in the 1.2 MDa complex of proteasome and BLM10 to form a largely closed dome that is expected to restrict access of potential substrates. Thus Blm10 and PA200 are predominantly nuclear and stimulate the degradation of model peptides, although they do not appear to stimulate the degradation of proteins, recognize ubiquitin, or utilize ATP. |
| 282094 | Mannosyl_trans2 | 1.78e-32 | 90 | 445 | 57 | 396 | Mannosyltransferase (PIG-V). This is a family of eukaryotic ER membrane proteins that are involved in the synthesis of glycosylphosphatidylinositol (GPI), a glycolipid that anchors many proteins to the eukaryotic cell surface. Proteins in this family are involved in transferring the second mannose in the biosynthetic pathway of GPI. |
| 406851 | BLM10_N | 3.10e-30 | 537 | 612 | 1 | 81 | Proteasome-substrate-size regulator, N-terminal. The ordered regions of the yeast BLM10 or PA200 (human homolog), full-length protein encode 32 HEAT repeat (HR)-like modules, each comprising two helices joined by a turn, with adjacent repeats connected by a linker. Whereas a standard HEAT repeat is composed of ~50 residues, the BLM10 HEAT repeats are highly variable. The length of helices ranges from 8 to 35 residues, turns range from 2 to 87 residues, and linkers range from 1 to 88 residues, with the longest linker, between HR21 and HR22, containing additional secondary structures (two strands and three helices). BLM10_N is the N-terminal ordered region of the three in BLM10. BLM10 is found to surround the proteasome entry pore in the 1.2 MDa complex of proteasome and BLM10 to form a largely closed dome that is expected to restrict access of potential substrates. BLM10 and PA200 are predominantly nuclear and stimulate the degradation of model peptides, although they do not appear to stimulate the degradation of proteins, recognize ubiquitin, or utilize ATP. |
| 227829 | COG5542 | 2.07e-12 | 92 | 444 | 76 | 367 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.13e-294 | 1 | 441 | 1 | 436 | |
| 1.20e-236 | 67 | 441 | 47 | 420 | |
| 7.38e-112 | 22 | 440 | 19 | 419 | |
| 1.61e-110 | 20 | 447 | 19 | 431 | |
| 6.32e-110 | 20 | 443 | 20 | 410 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.43e-71 | 575 | 2137 | 69 | 1548 | cryo-EM structure of human PA200 [Homo sapiens],6KWY_c human PA200-20S complex [Homo sapiens] |
|
| 2.37e-71 | 575 | 2137 | 34 | 1513 | Human 20S-PA200 Proteasome Complex [Homo sapiens],6REY_d Human 20S-PA200 Proteasome Complex [Homo sapiens] |
|
| 2.96e-39 | 615 | 1428 | 4 | 793 | Chain A5, Proteasome activator BLM10 [Saccharomyces cerevisiae],4V7O_A7 Chain A7, Proteasome activator BLM10 [Saccharomyces cerevisiae],4V7O_B4 Chain B4, Proteasome activator BLM10 [Saccharomyces cerevisiae],4V7O_B7 Chain B7, Proteasome activator BLM10 [Saccharomyces cerevisiae] |
|
| 5.60e-37 | 1490 | 2174 | 17 | 700 | Chain A6, Proteasome activator BLM10 [Saccharomyces cerevisiae],4V7O_A8 Chain A8, Proteasome activator BLM10 [Saccharomyces cerevisiae],4V7O_B5 Chain B5, Proteasome activator BLM10 [Saccharomyces cerevisiae],4V7O_B8 Chain B8, Proteasome activator BLM10 [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.53e-103 | 21 | 479 | 16 | 417 | GPI mannosyltransferase 2 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=gpi18 PE=3 SV=1 |
|
| 1.07e-95 | 2 | 444 | 3 | 399 | GPI mannosyltransferase 2 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=gpi18 PE=3 SV=1 |
|
| 1.28e-75 | 575 | 2137 | 19 | 1495 | Proteasome activator complex subunit 4B OS=Danio rerio OX=7955 GN=psme4b PE=3 SV=2 |
|
| 1.53e-74 | 544 | 2137 | 5 | 1513 | Proteasome activator complex subunit 4 OS=Mus musculus OX=10090 GN=Psme4 PE=1 SV=1 |
|
| 2.04e-74 | 615 | 2174 | 242 | 1846 | Proteasome activator BLM10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=BLM10 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.991758 | 0.008259 |
TMHMM Annotations download full data without filtering help
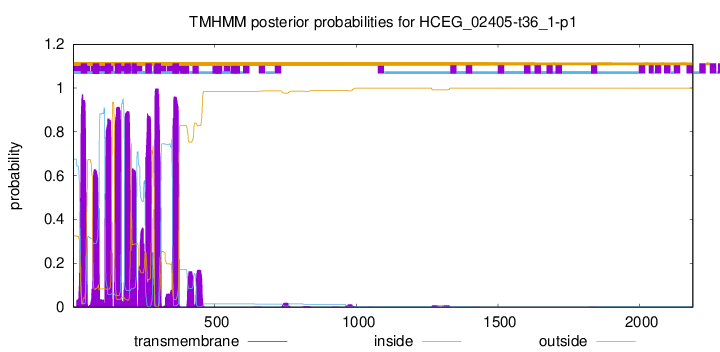
| Start | End |
|---|---|
| 27 | 49 |
| 69 | 91 |
| 112 | 134 |
| 147 | 169 |
| 181 | 203 |
| 207 | 224 |
| 255 | 277 |
| 287 | 309 |
| 351 | 373 |
