You are browsing environment: FUNGIDB
CAZyme Information: HCEG_00083-t36_1-p1
You are here: Home > Sequence: HCEG_00083-t36_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
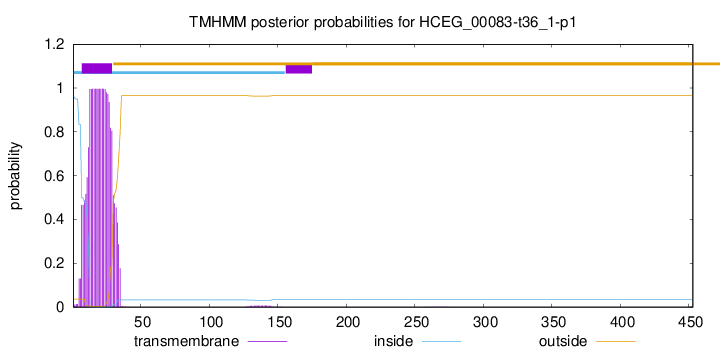
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | HCEG_00083-t36_1-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | DUF821 domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 310354 | Glyco_transf_90 | 8.56e-20 | 175 | 429 | 79 | 333 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
| 214773 | CAP10 | 5.54e-15 | 174 | 416 | 1 | 250 | Putative lipopolysaccharide-modifying enzyme. |
| 404418 | Glyco_trans_1_2 | 0.002 | 364 | 398 | 48 | 82 | Glycosyl transferases group 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 453 | 1 | 453 | |
| 0.0 | 1 | 453 | 1 | 453 | |
| 4.17e-166 | 75 | 446 | 68 | 439 | |
| 4.17e-166 | 75 | 446 | 68 | 439 | |
| 5.54e-163 | 68 | 446 | 57 | 436 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.39e-13 | 175 | 421 | 94 | 344 | human POGLUT1 in complex with Notch1 EGF12 and UDP [Homo sapiens],5L0S_A human POGLUT1 in complex with Factor VII EGF1 and UDP [Homo sapiens],5L0T_A human POGLUT1 in complex with EGF(+) and UDP [Homo sapiens],5L0U_A human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose [Homo sapiens],5L0V_A human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP [Homo sapiens],5UB5_A human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP [Homo sapiens] |
|
| 7.35e-10 | 175 | 422 | 128 | 382 | Crystal structure of Drosophila Poglut1 (Rumi) complexed with its glycoprotein product (glucosylated EGF repeat) and UDP [Drosophila melanogaster],5F85_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) and UDP [Drosophila melanogaster],5F86_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) [Drosophila melanogaster],5F87_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_B Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_C Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_D Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_E Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_F Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.97e-13 | 124 | 422 | 84 | 391 | O-glucosyltransferase rumi homolog OS=Aedes aegypti OX=7159 GN=AAEL011121 PE=3 SV=1 |
|
| 7.00e-13 | 59 | 422 | 15 | 392 | O-glucosyltransferase rumi homolog OS=Culex quinquefasciatus OX=7176 GN=CPIJ013394 PE=3 SV=1 |
|
| 8.85e-13 | 175 | 421 | 122 | 372 | Protein O-glucosyltransferase 1 OS=Homo sapiens OX=9606 GN=POGLUT1 PE=1 SV=1 |
|
| 6.67e-12 | 175 | 421 | 122 | 372 | Protein O-glucosyltransferase 1 OS=Bos taurus OX=9913 GN=POGLUT1 PE=2 SV=1 |
|
| 4.99e-11 | 175 | 421 | 122 | 372 | Protein O-glucosyltransferase 1 OS=Rattus norvegicus OX=10116 GN=Poglut1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
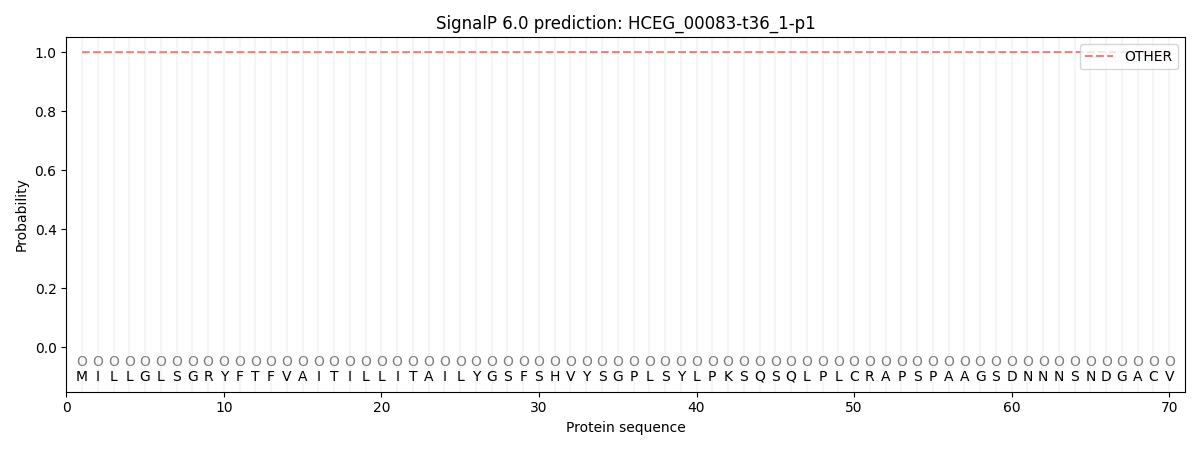
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999763 | 0.000257 |