You are browsing environment: FUNGIDB
CAZyme Information: HCDG_07930-t38_1-p1
You are here: Home > Sequence: HCDG_07930-t38_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | HCDG_07930-t38_1-p1 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 19 | 200 | 2.5e-71 | 0.9633507853403142 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185594 | PTZ00395 | 1.62e-06 | 230 | 276 | 417 | 463 | Sec24-related protein; Provisional |
| 223021 | PHA03247 | 2.68e-06 | 192 | 315 | 2739 | 2855 | large tegument protein UL36; Provisional |
| 185594 | PTZ00395 | 3.03e-06 | 230 | 317 | 432 | 517 | Sec24-related protein; Provisional |
| 185594 | PTZ00395 | 6.06e-06 | 96 | 312 | 327 | 545 | Sec24-related protein; Provisional |
| 237030 | kgd | 6.75e-06 | 232 | 311 | 35 | 114 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.61e-151 | 1 | 328 | 1 | 307 | |
| 1.89e-149 | 1 | 328 | 1 | 323 | |
| 2.70e-149 | 1 | 328 | 1 | 343 | |
| 5.10e-74 | 15 | 197 | 12 | 197 | |
| 1.57e-69 | 10 | 196 | 11 | 199 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.60e-39 | 19 | 195 | 1 | 196 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
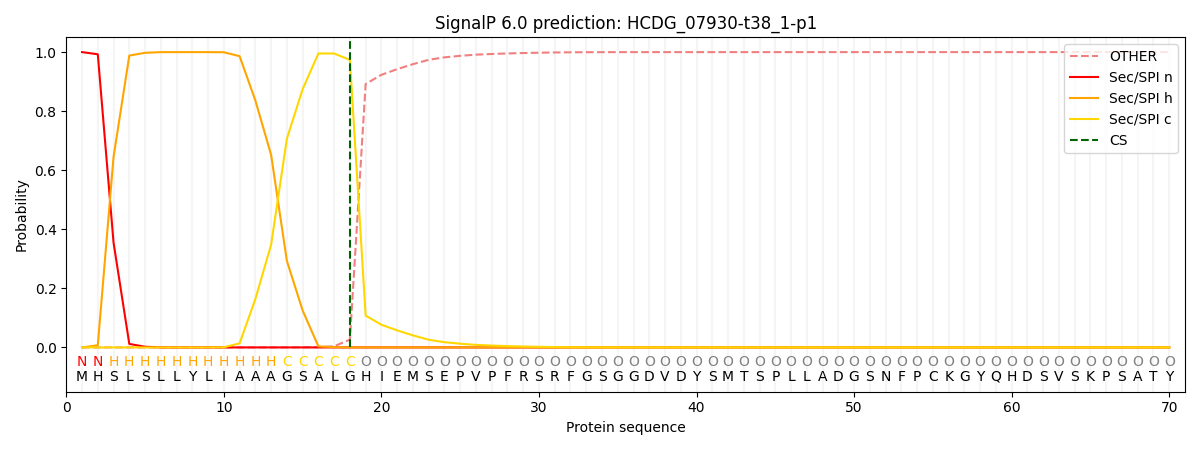
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000268 | 0.999688 | CS pos: 18-19. Pr: 0.9736 |
