You are browsing environment: FUNGIDB
CAZyme Information: HCDG_03729-t38_1-p1
You are here: Home > Sequence: HCDG_03729-t38_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | HCDG_03729-t38_1-p1 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | asparagine-linked glycosylation 9 protein isoform a | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.259:12 | 2.4.1.261:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 37 | 449 | 2.4e-97 | 0.961439588688946 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 1.82e-85 | 37 | 449 | 15 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 215437 | PLN02816 | 0.005 | 242 | 401 | 258 | 404 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 586 | 1 | 601 | |
| 0.0 | 1 | 586 | 1 | 602 | |
| 0.0 | 1 | 586 | 1 | 601 | |
| 1.40e-269 | 35 | 584 | 47 | 595 | |
| 5.16e-269 | 35 | 580 | 47 | 591 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.61e-119 | 41 | 577 | 34 | 557 | Alpha-1,2-mannosyltransferase alg9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg9 PE=3 SV=1 |
|
| 1.21e-101 | 35 | 560 | 72 | 584 | Alpha-1,2-mannosyltransferase ALG9 OS=Homo sapiens OX=9606 GN=ALG9 PE=1 SV=2 |
|
| 9.34e-101 | 35 | 560 | 72 | 584 | Alpha-1,2-mannosyltransferase ALG9 OS=Mus musculus OX=10090 GN=Alg9 PE=2 SV=1 |
|
| 1.15e-87 | 41 | 559 | 28 | 519 | Alpha-1,2-mannosyltransferase ALG9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG9 PE=1 SV=1 |
|
| 8.32e-77 | 41 | 579 | 66 | 567 | Dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG9 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000015 | 0.000003 |
TMHMM Annotations download full data without filtering help
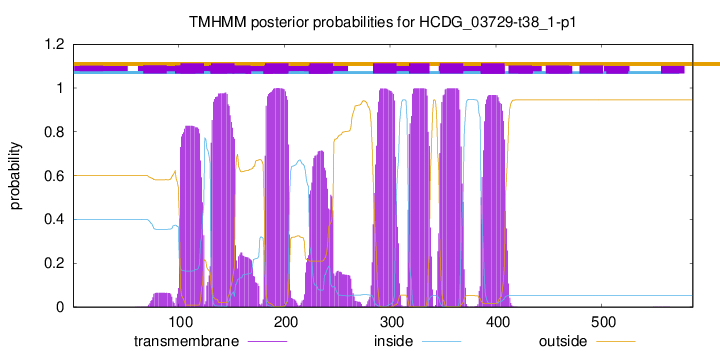
| Start | End |
|---|---|
| 102 | 124 |
| 131 | 153 |
| 182 | 204 |
| 224 | 246 |
| 284 | 306 |
| 318 | 337 |
| 347 | 369 |
| 386 | 408 |
