You are browsing environment: FUNGIDB
CAZyme Information: HCBG_02657-t26_1-p1
You are here: Home > Sequence: HCBG_02657-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | HCBG_02657-t26_1-p1 | |||||||||||
| CAZy Family | GH15|CBM20 | |||||||||||
| CAZyme Description | glucan synthase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:56 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 845 | 1623 | 0 | 0.9905277401894452 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 845 | 1661 | 1 | 819 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 7.17e-47 | 336 | 444 | 1 | 110 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 407550 | Trnau1ap | 5.16e-05 | 17 | 60 | 4 | 49 | Selenocysteine tRNA 1 associated proteins. This entry represents the C-terminal region of Selenocysteine tRNA 1 associated proteins (Trnau1ap also known as Secp43). Family members found in Eukaryotes have been shown to serve an essential role in the synthesis of selenoproteins, which have critical functions in numerous biological processes. Selenium deficiency results in a variety of diseases, including cardiac disease. Trnau1ap proteins harbor RNA recognition motifs (RRM) pfam00076 and Tyr-rich region found in the C-terminal. The Tyr-rich region (amino acids 185-225) is conserved among several mammals, including human, chimp, dog, cattle, mouse and rat. Furthermore, constitutive deletion of exons corresponding to the Tyr-rich region in mouse resulted in embryonic lethality. |
| 403477 | RCR | 0.001 | 6 | 73 | 42 | 105 | Chitin synthesis regulation, resistance to Congo red. RCR proteins are ER membrane proteins that regulate chitin deposition in fungal cell walls. Although chitin, a linear polymer of beta-1,4-linked N-acetylglucosamine, constitutes only 2% of the cell wall it plays a vital role in the overall protection of the cell wall against stress, noxious chemicals and osmotic pressure changes. Congo red is a cell wall-disrupting benzidine-type dye extensively used in many cell wall mutant studies that specifically targets chitin in yeast cells and inhibits growth. RCR proteins render the yeasts resistant to Congo red by diminishing the content of chitin in the cell wall. RCR proteins are probably regulating chitin synthase III interact directly with ubiquitin ligase Rsp5, and the VPEY motif is necessary for this, via interaction with the WW domains of Rsp5. |
| 407550 | Trnau1ap | 0.004 | 31 | 115 | 1 | 79 | Selenocysteine tRNA 1 associated proteins. This entry represents the C-terminal region of Selenocysteine tRNA 1 associated proteins (Trnau1ap also known as Secp43). Family members found in Eukaryotes have been shown to serve an essential role in the synthesis of selenoproteins, which have critical functions in numerous biological processes. Selenium deficiency results in a variety of diseases, including cardiac disease. Trnau1ap proteins harbor RNA recognition motifs (RRM) pfam00076 and Tyr-rich region found in the C-terminal. The Tyr-rich region (amino acids 185-225) is conserved among several mammals, including human, chimp, dog, cattle, mouse and rat. Furthermore, constitutive deletion of exons corresponding to the Tyr-rich region in mouse resulted in embryonic lethality. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1901 | 1 | 1901 | |
| 0.0 | 1 | 1901 | 1 | 1901 | |
| 0.0 | 1 | 1901 | 1 | 1901 | |
| 0.0 | 1 | 1884 | 1 | 1880 | |
| 0.0 | 79 | 1900 | 89 | 1902 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 185 | 1878 | 15 | 1763 | 1,3-beta-glucan synthase component FKS3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS3 PE=1 SV=1 |
|
| 0.0 | 71 | 1884 | 65 | 1878 | 1,3-beta-glucan synthase component FKS1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=fksA PE=3 SV=1 |
|
| 0.0 | 121 | 1880 | 82 | 1872 | 1,3-beta-glucan synthase component GSC2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GSC2 PE=1 SV=2 |
|
| 0.0 | 141 | 1848 | 110 | 1781 | 1,3-beta-glucan synthase component FKS1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=FKS1 PE=3 SV=3 |
|
| 0.0 | 102 | 1880 | 85 | 1853 | 1,3-beta-glucan synthase component FKS1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000025 | 0.000030 |
TMHMM Annotations download full data without filtering help
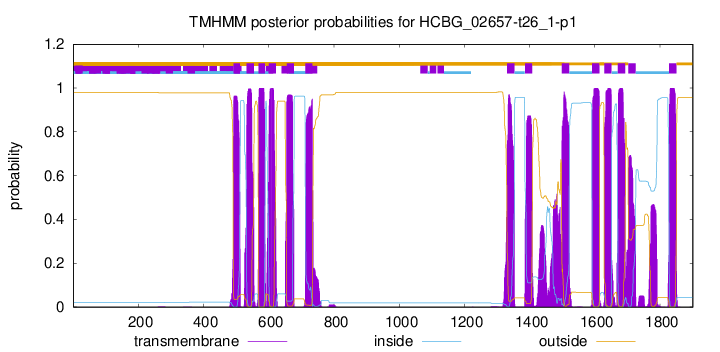
| Start | End |
|---|---|
| 492 | 511 |
| 532 | 554 |
| 569 | 588 |
| 600 | 622 |
| 655 | 677 |
| 712 | 734 |
| 1331 | 1353 |
| 1386 | 1408 |
| 1499 | 1521 |
| 1592 | 1614 |
| 1629 | 1651 |
| 1671 | 1693 |
| 1703 | 1725 |
| 1828 | 1850 |
