You are browsing environment: FUNGIDB
CAZyme Information: HCAG_06040-t26_1-p1
You are here: Home > Sequence: HCAG_06040-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | HCAG_06040-t26_1-p1 | |||||||||||
| CAZy Family | GT15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.52:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 188 | 424 | 4.1e-52 | 0.6735905044510386 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119332 | GH20_HexA_HexB-like | 7.56e-89 | 187 | 451 | 46 | 278 | Beta-N-acetylhexosaminidases catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. The hexA and hexB genes encode the alpha- and beta-subunits of the two major beta-N-acetylhexosaminidase isoenzymes, N-acetyl-beta-D-hexosaminidase A (HexA) and beta-N-acetylhexosaminidase B (HexB). Both the alpha and the beta catalytic subunits have a TIM-barrel fold and belong to the glycosyl hydrolase family 20 (GH20). The HexA enzyme is a heterodimer containing one alpha and one beta subunit while the HexB enzyme is a homodimer containing two beta-subunits. Hexosaminidase mutations cause an inability to properly hydrolyze certain sphingolipids which accumulate in lysosomes within the brain, resulting in the lipid storage disorders Tay-Sachs and Sandhoff. Mutations in the alpha subunit cause in a deficiency in the HexA enzyme and result in Tay-Sachs, mutations in the beta-subunit cause in a deficiency in both HexA and HexB enzymes and result in Sandhoff disease. In both disorders GM(2) gangliosides accumulate in lysosomes. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| 395590 | Glyco_hydro_20 | 2.21e-85 | 187 | 448 | 46 | 301 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| 373334 | Glycohydro_20b2 | 1.74e-41 | 28 | 162 | 1 | 133 | beta-acetyl hexosaminidase like. |
| 119333 | GH20_chitobiase-like | 8.12e-32 | 190 | 406 | 49 | 268 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| 119338 | GH20_chitobiase-like_1 | 9.88e-29 | 188 | 406 | 47 | 248 | A functionally uncharacterized subgroup of the Glycosyl hydrolase family 20 (GH20) catalytic domain found in proteins similar to the chitobiase of Serratia marcescens, a beta-N-1,4-acetylhexosaminidase that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This subgroup lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 453 | 1 | 481 | |
| 0.0 | 1 | 451 | 1 | 494 | |
| 0.0 | 1 | 451 | 1 | 494 | |
| 1.33e-181 | 16 | 461 | 14 | 506 | |
| 1.27e-180 | 23 | 451 | 18 | 492 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.00e-158 | 109 | 451 | 5 | 391 | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae [Aspergillus oryzae],5OAR_D Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae [Aspergillus oryzae] |
|
| 3.47e-41 | 109 | 451 | 109 | 478 | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis [Ostrinia furnacalis],3NSN_A Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin [Ostrinia furnacalis],3OZO_A Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT [Ostrinia furnacalis],3OZP_A Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with PUGNAc [Ostrinia furnacalis] |
|
| 6.55e-41 | 109 | 451 | 109 | 478 | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 [Ostrinia furnacalis],3WMC_A Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 [Ostrinia furnacalis] |
|
| 7.03e-41 | 109 | 451 | 109 | 478 | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with berberine [Ostrinia furnacalis] |
|
| 7.03e-41 | 109 | 451 | 109 | 478 | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with a berberine derivative (SYSU-00679) [Ostrinia furnacalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.35e-182 | 16 | 461 | 14 | 506 | Beta-hexosaminidase 1 OS=Coccidioides posadasii (strain RMSCC 757 / Silveira) OX=443226 GN=HEX1 PE=1 SV=1 |
|
| 2.25e-181 | 23 | 451 | 18 | 492 | Beta-hexosaminidase OS=Aspergillus oryzae OX=5062 GN=nagA PE=1 SV=1 |
|
| 4.62e-179 | 21 | 451 | 17 | 495 | Beta-hexosaminidase OS=Emericella nidulans OX=162425 GN=nagA PE=1 SV=1 |
|
| 5.91e-154 | 11 | 451 | 20 | 507 | Probable beta-hexosaminidase ARB_01353 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_01353 PE=1 SV=1 |
|
| 6.01e-95 | 16 | 451 | 15 | 455 | Beta-hexosaminidase OS=Candida albicans OX=5476 GN=HEX1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
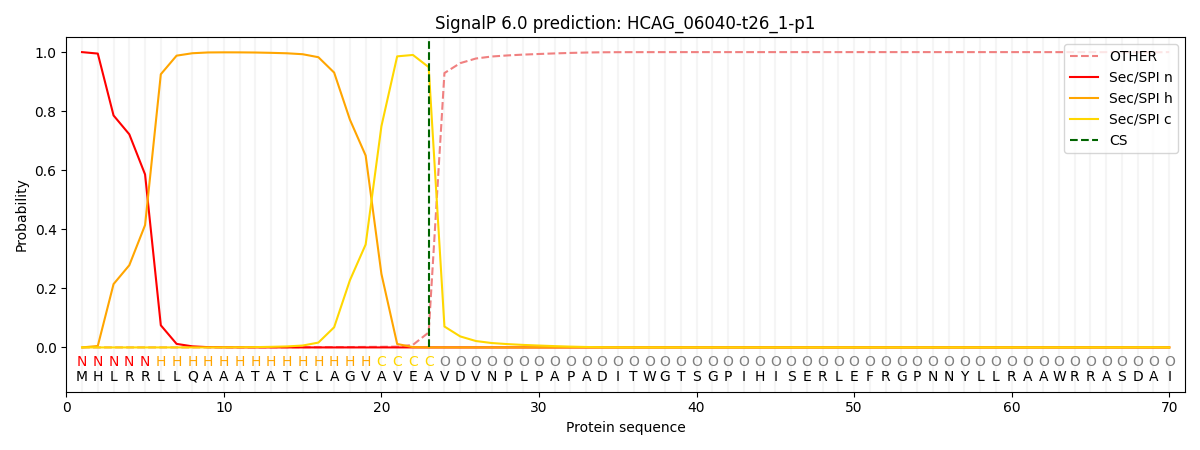
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000909 | 0.999067 | CS pos: 23-24. Pr: 0.9491 |
