You are browsing environment: FUNGIDB
CAZyme Information: H310_14500-t26_1-p1
Basic Information
help
| Species |
Aphanomyces invadans
|
| Lineage |
Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans
|
| CAZyme ID |
H310_14500-t26_1-p1
|
| CAZy Family |
GT71 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 922 |
KI914023|CGC1 |
100183.58 |
5.2325 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AinvadansNJM9701 |
15416 |
N/A |
168 |
15248
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH17 |
390 |
592 |
3.7e-16 |
0.7427652733118971 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227625
|
Scw11 |
3.94e-15 |
319 |
592 |
47 |
303 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 227625
|
Scw11 |
1.02e-12 |
51 |
278 |
67 |
285 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 227625
|
Scw11 |
9.43e-10 |
638 |
909 |
44 |
305 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.24e-232 |
8 |
919 |
2 |
908 |
| 6.73e-169 |
21 |
565 |
20 |
559 |
| 2.39e-98 |
301 |
590 |
7 |
301 |
| 6.20e-93 |
298 |
606 |
77 |
382 |
| 8.04e-93 |
305 |
604 |
10 |
317 |
H310_14500-t26_1-p1 has no PDB hit.
H310_14500-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
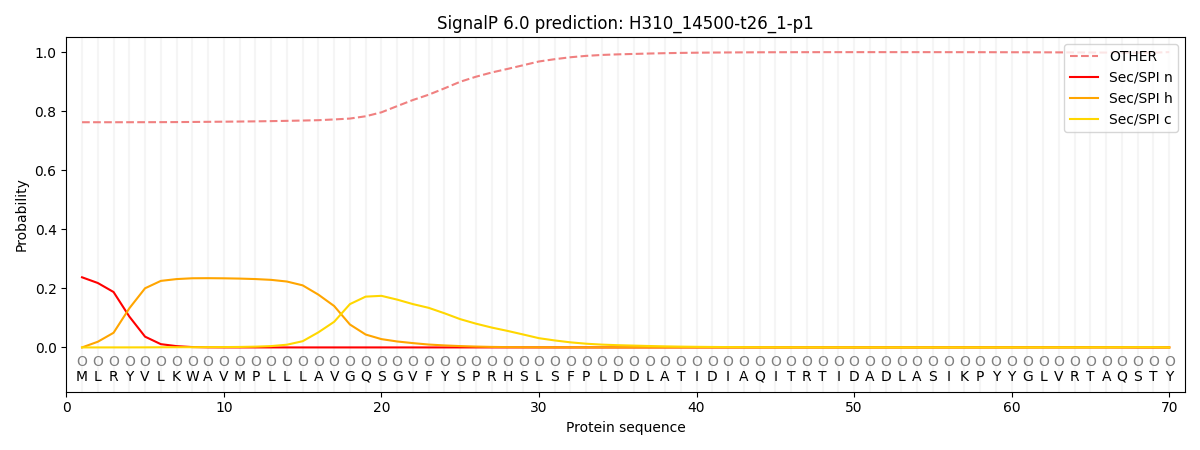
| Other |
SP_Sec_SPI |
CS Position |
| 0.774890 |
0.225119 |
|
There is no transmembrane helices in H310_14500-t26_1-p1.

