You are browsing environment: FUNGIDB
CAZyme Information: H310_14114-t26_1-p1
You are here: Home > Sequence: H310_14114-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_14114-t26_1-p1 | |||||||||||
| CAZy Family | GT60 | |||||||||||
| CAZyme Description | hypothetical protein, variant 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 264 | 907 | 4.5e-60 | 0.5205673758865248 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 7.60e-29 | 543 | 903 | 259 | 588 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 239042 | PA_hPAP21_like | 8.97e-06 | 52 | 126 | 1 | 70 | PA_hPAP21_like: Protease-associated domain containing proteins like the human secreted glycoprotein hPAP21 (human protease-associated domain-containing protein, 21kDa). This group contains various PA domain-containing proteins similar to hPAP21. Complex N-glycosylation may be required for the secretion of hPAP21. The significance of the PA domain to hPAP21 has not been ascertained. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. |
| 223515 | RfaB | 1.89e-04 | 804 | 913 | 258 | 363 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| 239041 | PA_EDEM3_like | 2.18e-04 | 65 | 126 | 20 | 77 | PA_EDEM3_like: protease associated domain (PA) domain-containing EDEM3-like proteins. This group contains various PA domain-containing proteins similar to mouse EDEM3 (ER-degradation-enhancing mannosidase-like 3 protein). EDEM3 contains a region, similar to Class I alpha-mannosidases (gylcosyl hydrolase family 47), N-terminal to the PA domain. EDEM3 accelerates glycoprotein ERAD (ER-associated degradation). In transfected mammalian cells, overexpression of EDEM3 enhances the mannose trimming from the N-glycans, of a model misfolded protein [alpha1-antitrypsin null (Hong Kong)] as well as, from total glycoproteins. Mannose trimming appears to be involved in the selection of ERAD substrates. EDEM3 has a different specificity of trimming than ER alpha-mannosidase 1. The significance of the PA domain to EDEM3 has not been ascertained. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. |
| 340831 | GT4_PimA-like | 3.86e-04 | 545 | 913 | 1 | 354 | phosphatidyl-myo-inositol mannosyltransferase. This family is most closely related to the GT4 family of glycosyltransferases and named after PimA in Propionibacterium freudenreichii, which is involved in the biosynthesis of phosphatidyl-myo-inositol mannosides (PIM) which are early precursors in the biosynthesis of lipomannans (LM) and lipoarabinomannans (LAM), and catalyzes the addition of a mannosyl residue from GDP-D-mannose (GDP-Man) to the position 2 of the carrier lipid phosphatidyl-myo-inositol (PI) to generate a phosphatidyl-myo-inositol bearing an alpha-1,2-linked mannose residue (PIM1). Glycosyltransferases catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. The acceptor molecule can be a lipid, a protein, a heterocyclic compound, or another carbohydrate residue. This group of glycosyltransferases is most closely related to the previously defined glycosyltransferase family 1 (GT1). The members of this family may transfer UDP, ADP, GDP, or CMP linked sugars. The diverse enzymatic activities among members of this family reflect a wide range of biological functions. The protein structure available for this family has the GTB topology, one of the two protein topologies observed for nucleotide-sugar-dependent glycosyltransferases. GTB proteins have distinct N- and C- terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. The members of this family are found mainly in certain bacteria and archaea. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.14e-205 | 22 | 928 | 22 | 874 | |
| 8.79e-65 | 432 | 903 | 508 | 926 | |
| 3.25e-63 | 437 | 903 | 511 | 924 | |
| 2.83e-59 | 431 | 912 | 236 | 669 | |
| 4.64e-59 | 404 | 912 | 379 | 838 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.41e-13 | 635 | 903 | 356 | 599 | Crystal structure of the Actinobacillus pleuropneumoniae HMW1C glycosyltransferase [Actinobacillus pleuropneumoniae serovar 1 str. 4074],3Q3E_B Crystal structure of the Actinobacillus pleuropneumoniae HMW1C glycosyltransferase [Actinobacillus pleuropneumoniae serovar 1 str. 4074],3Q3H_A Crystal structure of the Actinobacillus pleuropneumoniae HMW1C glycosyltransferase in complex with UDP-GLC [Actinobacillus pleuropneumoniae serovar 1 str. 4074],3Q3H_B Crystal structure of the Actinobacillus pleuropneumoniae HMW1C glycosyltransferase in complex with UDP-GLC [Actinobacillus pleuropneumoniae serovar 1 str. 4074],3Q3I_A Crystal structure of the Actinobacillus pleuropneumoniae HMW1C glycosyltransferase in the presence of peptide N1131 [Actinobacillus pleuropneumoniae serovar 1 str. 4074],3Q3I_B Crystal structure of the Actinobacillus pleuropneumoniae HMW1C glycosyltransferase in the presence of peptide N1131 [Actinobacillus pleuropneumoniae serovar 1 str. 4074] |
|
| 3.20e-12 | 508 | 889 | 113 | 475 | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_B Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_C Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_D Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum] |
|
| 7.24e-11 | 544 | 903 | 206 | 533 | Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2JLB_B Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2VSY_A Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2VSY_B Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2XGM_A Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGM_B Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGO_A XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGO_B XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGS_A XcOGT in complex with C-UDP [Xanthomonas campestris],2XGS_B XcOGT in complex with C-UDP [Xanthomonas campestris] |
|
| 9.54e-11 | 544 | 903 | 206 | 533 | Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004],2VSN_B Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.79e-13 | 635 | 903 | 345 | 588 | UDP-glucose:protein N-beta-glucosyltransferase OS=Actinobacillus pleuropneumoniae serotype 7 (strain AP76) OX=537457 GN=APP7_1697 PE=1 SV=1 |
|
| 7.12e-13 | 635 | 903 | 345 | 588 | UDP-glucose:protein N-beta-glucosyltransferase OS=Actinobacillus pleuropneumoniae serotype 5b (strain L20) OX=416269 GN=APL_1635 PE=1 SV=1 |
|
| 2.02e-09 | 543 | 903 | 591 | 925 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC OS=Arabidopsis thaliana OX=3702 GN=SEC PE=1 SV=1 |
|
| 3.40e-09 | 537 | 905 | 476 | 816 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY OS=Arabidopsis thaliana OX=3702 GN=SPY PE=1 SV=1 |
|
| 1.18e-07 | 541 | 905 | 485 | 821 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY OS=Petunia hybrida OX=4102 GN=SPY PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
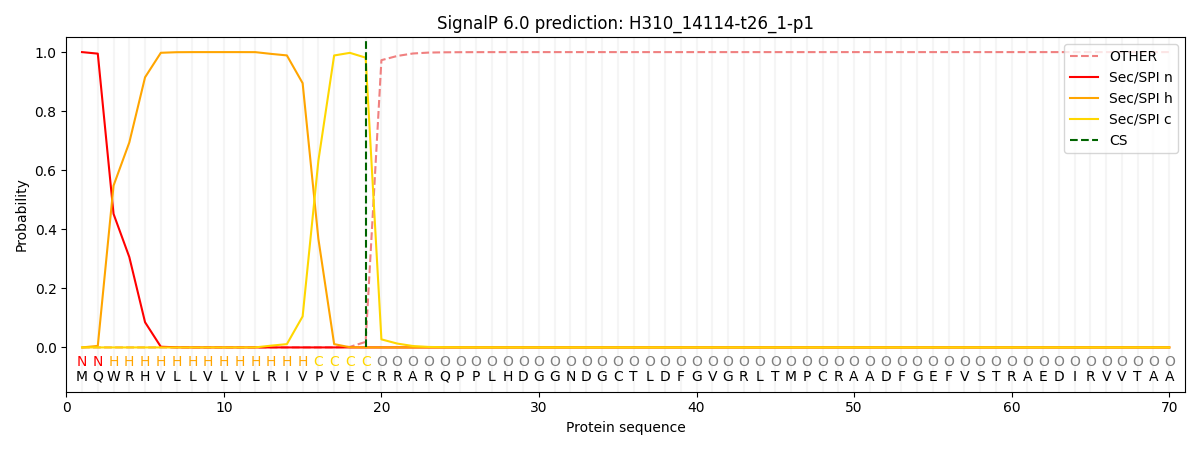
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000205 | 0.999781 | CS pos: 19-20. Pr: 0.9812 |
