You are browsing environment: FUNGIDB
CAZyme Information: H310_11320-t26_1-p1
You are here: Home > Sequence: H310_11320-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_11320-t26_1-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 183 | 300 | 4.9e-21 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395611 | F5_F8_type_C | 2.19e-19 | 181 | 298 | 2 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| 225201 | WD40 | 3.35e-10 | 1383 | 1704 | 160 | 458 | WD40 repeat [General function prediction only]. |
| 293791 | 7WD40 | 4.50e-09 | 1669 | 1952 | 9 | 288 | WD40 repeats in seven bladed beta propellers. The WD40 repeat is found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing, and cytoskeleton assembly. It typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40. Between the GH and WD dipeptides lies a conserved core. It forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel beta-sheet. The WD40 sequence repeat originally described in literature forms the first three strands of one blade and the last strand in the next blade. The C-terminal WD40 repeat completes the blade structure of the N-terminal WD40 repeat to create the closed ring propeller-structure. The residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands, allowing them to bind either stably or reversibly. |
| 404200 | DUF4062 | 2.09e-08 | 499 | 585 | 2 | 68 | Domain of unknown function (DUF4062). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 80 amino acids in length. There is a conserved SST sequence motif. |
| 225201 | WD40 | 7.31e-08 | 1606 | 1954 | 57 | 436 | WD40 repeat [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.47e-24 | 175 | 317 | 53 | 190 | |
| 8.07e-24 | 175 | 308 | 37 | 166 | |
| 8.07e-24 | 175 | 308 | 37 | 166 | |
| 8.07e-24 | 175 | 308 | 37 | 166 | |
| 1.89e-23 | 175 | 304 | 36 | 160 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.35e-21 | 174 | 303 | 7 | 136 | Solution structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
|
| 3.45e-21 | 174 | 303 | 8 | 137 | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
|
| 6.41e-21 | 174 | 303 | 8 | 137 | X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
|
| 6.71e-19 | 174 | 303 | 7 | 137 | Solution structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
|
| 6.90e-19 | 174 | 303 | 8 | 138 | X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ5_B X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_A X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_B X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.78e-74 | 531 | 1285 | 23 | 771 | NACHT domain- and WD repeat-containing protein 1 OS=Homo sapiens OX=9606 GN=NWD1 PE=1 SV=3 |
|
| 1.78e-72 | 510 | 1254 | 3 | 739 | NACHT domain- and WD repeat-containing protein 1 OS=Mus musculus OX=10090 GN=Nwd1 PE=2 SV=2 |
|
| 1.38e-67 | 473 | 1197 | 15 | 763 | NACHT and WD repeat domain-containing protein 2 OS=Mus musculus OX=10090 GN=Nwd2 PE=1 SV=2 |
|
| 7.10e-67 | 473 | 1197 | 15 | 763 | NACHT and WD repeat domain-containing protein 2 OS=Homo sapiens OX=9606 GN=NWD2 PE=2 SV=3 |
|
| 4.17e-17 | 497 | 1116 | 912 | 1449 | Telomerase protein component 1 OS=Rattus norvegicus OX=10116 GN=Tep1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
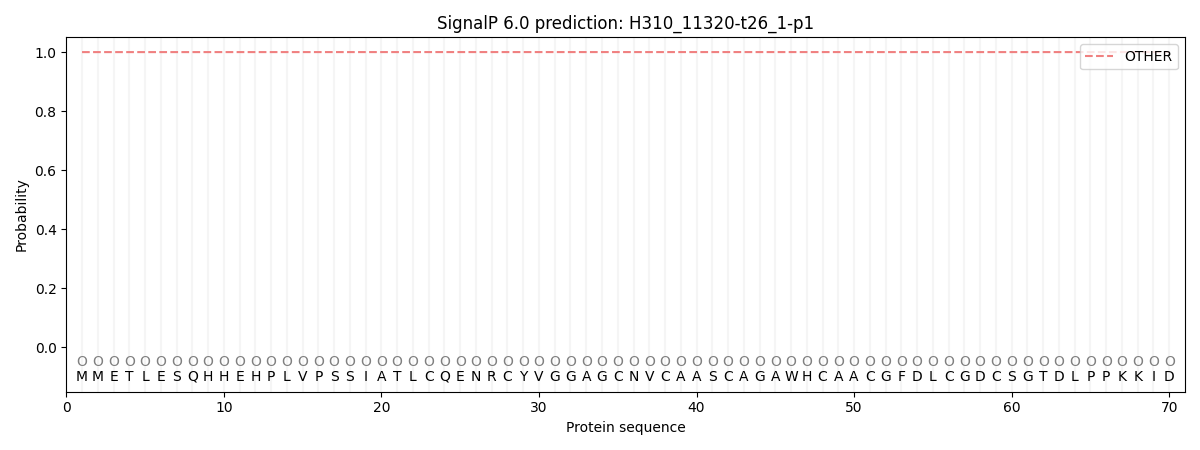
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000051 | 0.000001 |
