You are browsing environment: FUNGIDB
CAZyme Information: H310_11285-t26_1-p1
You are here: Home > Sequence: H310_11285-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
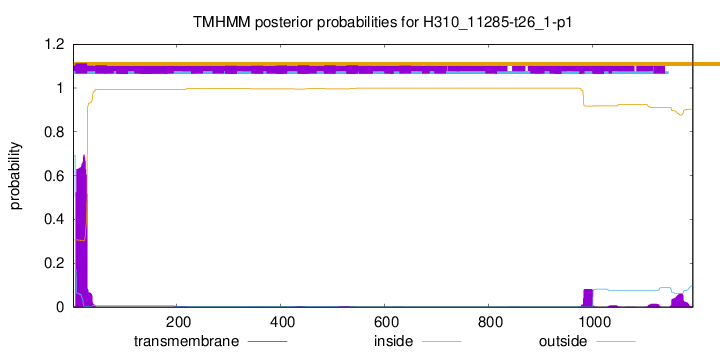
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_11285-t26_1-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 547317; End:551399 Strand: + | |||||||||||
Full Sequence Download help
| MGRHVAWVSM LGIQKMAMLA IAAHALMNGS GLLSGAVEEF RVSRGMIDFR GDIGQIGRTT | 60 |
| STIKQQSQRS SYIVSMKSMH RFCKDLSEAL DHDTAERCWV TLFDTFPNVD RSPMAFQVHY | 120 |
| DYARHIENGS NDRTSAAKTA YERALAVMTE PSSPLSLDAM NRLGLLWQKL SRPHVAIEVF | 180 |
| RQSLQWHPHE CALRGNLASA YLELGNVAAA LAAAAVALKL EPHNAALHHN MGNIYQRAGD | 240 |
| MLSAMSHWQK AVEIDPQNPN PLFSLGIAAG NQGLSDTSTA YFAKSVVVAS QHAPALRDLA | 300 |
| RLQLATALLP RVYDSTNDIH QIRQTFTTAL EDLMADSPLS LGDNPIYTTG CGSMGYYLVY | 360 |
| QGGPNKHLRR TLAQIYEKSM PGLEFEAPHV RAARHHTFQF PSTARFVDAR PSNVSAAVIV | 420 |
| HRRIRVGILS AFMYHHSVGL LMQGVLTRLS NATYELVLLR CRPWHADHVT DRVSRHMSRV | 480 |
| VQLPNDVFHA QQVVADLALD ILLFTELGMD ASSYFLAFAT LALRSVLFWG HAVTSGLSTV | 540 |
| DYFVTSTHFQ SNPVSGSSDE FTETLYPMQS LTTVFHRPRF SFHRSLLSFL PADDDGRVMY | 600 |
| LIPQTLYKLH PAMDSLVAAI LAGHRNSYLV LLEGAVPHLA TQIRRRLGRV LSLQDLESRV | 660 |
| VFAPYLSTDA FLTLCQVADV VLDPFPVGGG RSSFEVFSVG TPIVAHTART SIAQLTAAMY | 720 |
| TKMNITACCV AATDEAYETL DSSVATALAL GQNTSYRQEM STLIQSRSSV LFESDASTVA | 780 |
| SVMDDWNRCF HTILAMPPPP RPTPASTVAI PFGFRLQLFG QLSVEVVMHA NETPTQVGAT | 840 |
| IAALYGLEPL YQCYIQTLLF HGVQRWRQNE AQLAITTDAF PGRTPPLDIY VGDDVALAVQ | 900 |
| YHLWRYNHRV PGRDVAVAVA SIRHKLAFSD ASAPWIAARG VQHRPPRSAV QAHDAPLQST | 960 |
| RQCLTVALTT CKRLSLFLAT MKSFLPYANS TAVAICMILV LDDHSSAADR RVMQESFPTV | 1020 |
| QFVFTRRKGH AHSLNLLLDL VSTPFVLYLE DDWQWLQDLP HHPIAHALAV LDSDPTLVQV | 1080 |
| LLNTQFSGWP RTMHDSHPFD VYFRHEFGIV DHAFGYWPGF SLNPGVWNLR RLRQCPRLTF | 1140 |
| NERSDVFERE FSLAVWQCGL HVAMLPYTTA VHIGAAPGTN GSAYALNGMP RRFD | 1194 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 129 | 772 | 2.1e-67 | 0.6226950354609929 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 1.15e-26 | 135 | 776 | 1 | 602 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 276809 | TPR | 6.30e-12 | 158 | 253 | 2 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 274350 | PEP_TPR_lipo | 9.83e-11 | 137 | 307 | 375 | 547 | putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. |
| 276809 | TPR | 3.70e-10 | 196 | 284 | 6 | 94 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 1.66e-09 | 225 | 305 | 1 | 81 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBO34148.1|GT41 | 1.02e-52 | 304 | 776 | 516 | 938 |
| QGJ68883.1|GT41 | 1.31e-50 | 304 | 788 | 280 | 720 |
| BAP54338.1|GT41 | 1.77e-49 | 301 | 773 | 138 | 570 |
| BBO66743.1|GT41 | 4.88e-47 | 308 | 773 | 463 | 886 |
| ACB77232.1|GT41 | 5.67e-47 | 309 | 740 | 348 | 748 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5DJS_A | 1.05e-06 | 421 | 704 | 160 | 436 | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_B Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_C Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_D Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum] |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
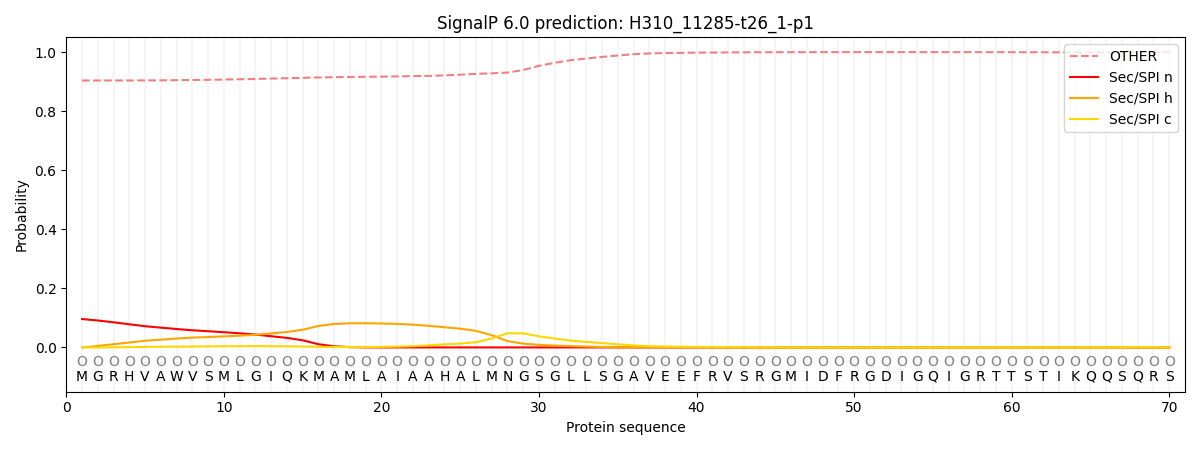
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.910466 | 0.089543 |