You are browsing environment: FUNGIDB
CAZyme Information: H310_11182-t26_1-p1
You are here: Home > Sequence: H310_11182-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_11182-t26_1-p1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 864 | 1598 | 1.9e-268 | 0.9499323410013532 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 865 | 1546 | 3 | 701 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 1.45e-28 | 171 | 262 | 8 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 395036 | Sugar_tr | 1.28e-13 | 1810 | 2171 | 58 | 428 | Sugar (and other) transporter. |
| 340917 | MFS_XylE_like | 1.25e-06 | 1783 | 2166 | 21 | 377 | D-xylose-proton symporter and similar transporters of the Major Facilitator Superfamily. This subfamily includes bacterial transporters such as D-xylose-proton symporter (XylE or XylT), arabinose-proton symporter (AraE), galactose-proton symporter (GalP), major myo-inositol transporter IolT, glucose transport protein, putative metabolite transport proteins YfiG, YncC, and YwtG, and similar proteins. The symporters XylE, AraE, and GalP facilitate the uptake of D-xylose, arabinose, and galactose, respectively, across the boundary membrane with the concomitant transport of protons into the cell. IolT is involved in polyol metabolism and myo-inositol degradation into acetyl-CoA. The XylE-like subfamily belongs to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 369468 | MFS_1 | 2.48e-06 | 1775 | 2106 | 5 | 318 | Major Facilitator Superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 2189 | 9 | 2183 | |
| 0.0 | 4 | 2222 | 15 | 2216 | |
| 0.0 | 5 | 2255 | 10 | 2242 | |
| 0.0 | 4 | 2219 | 13 | 2212 | |
| 0.0 | 4 | 2204 | 12 | 2188 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.25e-236 | 94 | 1751 | 264 | 1918 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 2.45e-226 | 88 | 1751 | 265 | 1892 | Callose synthase 10 OS=Arabidopsis thaliana OX=3702 GN=CALS10 PE=2 SV=5 |
|
| 4.83e-226 | 76 | 1751 | 240 | 1878 | Callose synthase 9 OS=Arabidopsis thaliana OX=3702 GN=CALS9 PE=2 SV=2 |
|
| 4.67e-225 | 90 | 1751 | 251 | 1906 | Putative callose synthase 6 OS=Arabidopsis thaliana OX=3702 GN=CALS6 PE=3 SV=2 |
|
| 5.83e-225 | 88 | 1746 | 77 | 1763 | Callose synthase 12 OS=Arabidopsis thaliana OX=3702 GN=CALS12 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
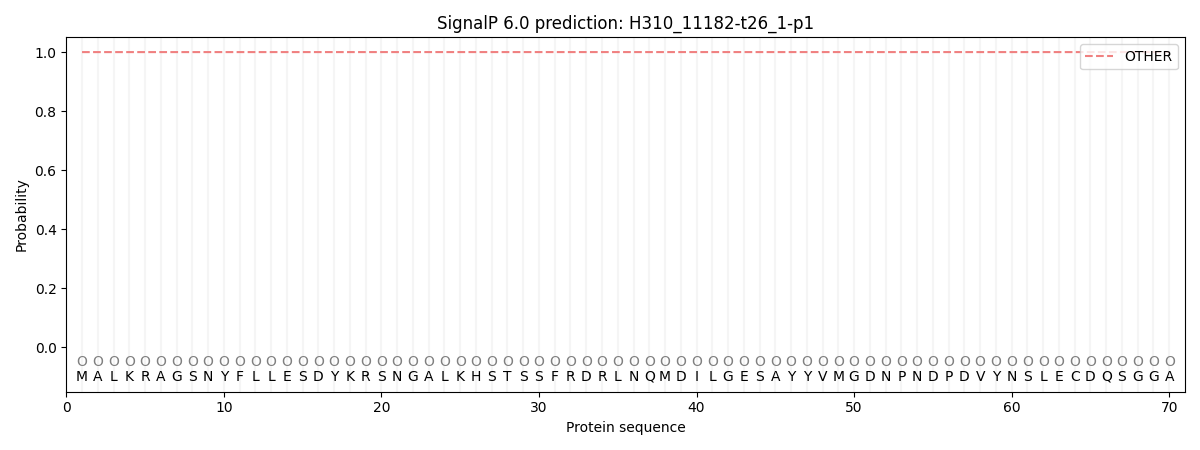
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000001 |
TMHMM Annotations download full data without filtering help
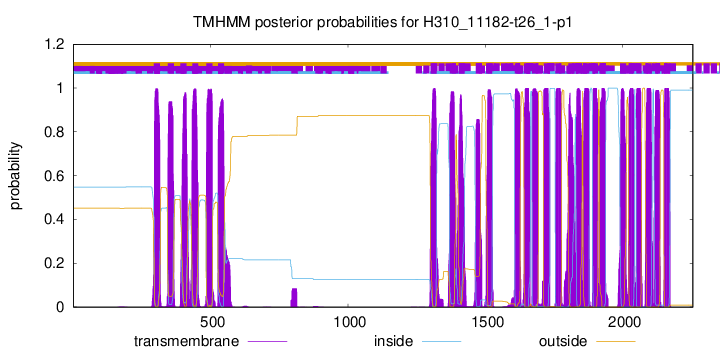
| Start | End |
|---|---|
| 293 | 315 |
| 344 | 366 |
| 390 | 412 |
| 433 | 455 |
| 484 | 506 |
| 527 | 549 |
| 1300 | 1322 |
| 1370 | 1392 |
| 1402 | 1424 |
| 1465 | 1487 |
| 1502 | 1524 |
| 1607 | 1626 |
| 1641 | 1663 |
| 1670 | 1692 |
| 1712 | 1734 |
| 1754 | 1776 |
| 1802 | 1824 |
| 1829 | 1851 |
| 1855 | 1877 |
| 1890 | 1909 |
| 1915 | 1937 |
| 1990 | 2012 |
| 2022 | 2042 |
| 2049 | 2068 |
| 2083 | 2105 |
| 2118 | 2135 |
| 2150 | 2172 |
