You are browsing environment: FUNGIDB
CAZyme Information: H310_09882-t26_1-p1
You are here: Home > Sequence: H310_09882-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_09882-t26_1-p1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 865 | 1621 | 1.7e-273 | 0.972936400541272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 866 | 1552 | 3 | 701 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 5.09e-26 | 171 | 269 | 8 | 109 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 340916 | MFS_GLUT6_8_Class3_like | 9.22e-23 | 1771 | 2191 | 5 | 424 | Glucose transporter (GLUT) types 6 and 8, Class 3 GLUTs, and similar transporters of the Major Facilitator Superfamily. This subfamily is composed of glucose transporter type 6 (GLUT6), GLUT8, plant early dehydration-induced gene ERD6-like proteins, and similar insect proteins including facilitated trehalose transporter Tret1-1. GLUTs, also called Solute carrier family 2, facilitated glucose transporters (SLC2A), are a family of proteins that facilitate the transport of hexoses such as glucose and fructose. There are fourteen GLUTs found in humans; they display different substrate specificities and tissue expression. They have been categorized into three classes based on sequence similarity: Class 1 (GLUTs 1-4, 14); Class 2 (GLUTs 5, 7, 9, and 11); and Class 3 (GLUTs 6, 8, 10, 12, and HMIT). Insect Tret1-1 is a low-capacity facilitative transporter for trehalose that mediates the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source. GLUT proteins are comprised of about 500 amino acid residues, possess a single N-linked oligosaccharide, and have 12 transmembrane segments. They belong to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 395036 | Sugar_tr | 5.08e-21 | 1770 | 2144 | 2 | 387 | Sugar (and other) transporter. |
| 340915 | MFS_GLUT_Class1_2_like | 1.58e-14 | 1825 | 2123 | 61 | 373 | Class 1 and Class 2 Glucose transporters (GLUTs) of the Major Facilitator Superfamily. This subfamily includes Class 1 and Class 2 glucose transporters (GLUTs) including Solute carrier family 2, facilitated glucose transporter member 1 (SLC2A1, also called glucose transporter type 1 or GLUT1), SLC2A2-5 (GLUT2-5), SLC2A7 (GLUT7), SLC2A9 (GLUT9), SLC2A11 (GLUT11), SLC2A14 (GLUT14), and similar proteins. GLUTs are a family of proteins that facilitate the transport of hexoses such as glucose and fructose. There are fourteen GLUTs found in humans; they display different substrate specificities and tissue expression. They have been categorized into three classes based on sequence similarity: Class 1 (GLUTs 1-4, 14); Class 2 (GLUTs 5, 7, 9, and 11); and Class 3 (GLUTs 6, 8, 10, 12, and HMIT). GLUTs 1-5 are the most thoroughly studied and are well-established as glucose and/or fructose transporters in various tissues and cell types. GLUT proteins are comprised of about 500 amino acid residues, possess a single N-linked oligosaccharide, and have 12 transmembrane segments. They belong to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 19 | 2264 | 6 | 2238 | |
| 0.0 | 16 | 2264 | 5 | 2224 | |
| 0.0 | 22 | 2260 | 1 | 2240 | |
| 0.0 | 24 | 2264 | 6 | 2222 | |
| 0.0 | 25 | 2262 | 8 | 2232 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.27e-233 | 84 | 1764 | 258 | 1918 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 1.35e-229 | 86 | 1759 | 244 | 1908 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 6.13e-227 | 84 | 1764 | 253 | 1878 | Callose synthase 9 OS=Arabidopsis thaliana OX=3702 GN=CALS9 PE=2 SV=2 |
|
| 1.04e-226 | 90 | 1759 | 83 | 1763 | Callose synthase 12 OS=Arabidopsis thaliana OX=3702 GN=CALS12 PE=2 SV=1 |
|
| 7.16e-223 | 84 | 1764 | 265 | 1892 | Callose synthase 10 OS=Arabidopsis thaliana OX=3702 GN=CALS10 PE=2 SV=5 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000049 | 0.000000 |
TMHMM Annotations download full data without filtering help
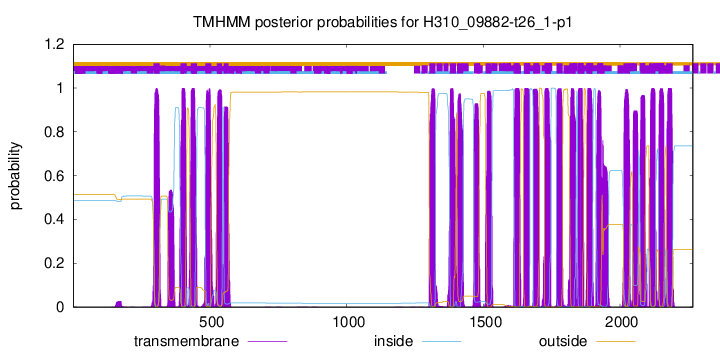
| Start | End |
|---|---|
| 294 | 316 |
| 393 | 415 |
| 430 | 452 |
| 483 | 505 |
| 525 | 544 |
| 551 | 568 |
| 1303 | 1325 |
| 1378 | 1400 |
| 1404 | 1426 |
| 1466 | 1488 |
| 1508 | 1530 |
| 1611 | 1633 |
| 1648 | 1670 |
| 1683 | 1705 |
| 1725 | 1747 |
| 1768 | 1790 |
| 1816 | 1838 |
| 1845 | 1867 |
| 1877 | 1899 |
| 1916 | 1938 |
| 2014 | 2036 |
| 2075 | 2092 |
| 2107 | 2129 |
| 2142 | 2161 |
| 2171 | 2193 |
