You are browsing environment: FUNGIDB
CAZyme Information: H310_09456-t26_1-p1
You are here: Home > Sequence: H310_09456-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_09456-t26_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT30 | 376 | 545 | 2.6e-48 | 0.9096045197740112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235589 | PRK05749 | 4.21e-114 | 376 | 724 | 51 | 391 | 3-deoxy-D-manno-octulosonic-acid transferase; Reviewed |
| 224436 | KdtA | 9.21e-111 | 376 | 724 | 50 | 389 | 3-deoxy-D-manno-octulosonic-acid transferase [Cell wall/membrane/envelope biogenesis]. |
| 398219 | Glycos_transf_N | 2.69e-57 | 376 | 544 | 14 | 174 | 3-Deoxy-D-manno-octulosonic-acid transferase (kdotransferase). Members of this family transfer activated sugars to a variety of substrates, including glycogen, fructose-6-phosphate and lipopolysaccharides. Members of the family transfer UDP, ADP, GDP or CMP linked sugars. The Glycos_transf_N region is flanked at the N-terminus by a signal peptide and at the C-terminus by Glycos_transf_1 (pfam00534). The eukaryotic glycogen synthases may be distant members of this bacterial family. |
| 395687 | SET | 2.88e-14 | 20 | 118 | 3 | 114 | SET domain. SET domains are protein lysine methyltransferase enzymes. SET domains appear to be protein-protein interaction domains. It has been demonstrated that SET domains mediate interactions with a family of proteins that display similarity with dual-specificity phosphatases (dsPTPases). A subset of SET domains have been called PR domains. These domains are divergent in sequence from other SET domains, but also appear to mediate protein-protein interaction. The SET domain consists of two regions known as SET-N and SET-C. SET-C forms an unusual and conserved knot-like structure of probably functional importance. Additionally to SET-N and SET-C, an insert region (SET-I) and flanking regions of high structural variability form part of the overall structure. |
| 380929 | SET_SETD2-like | 1.19e-13 | 190 | 318 | 12 | 136 | SET domain (including post-SET domain) found in SET domain-containing protein 2 (SETD2), nuclear SETD2 (NSD2), ASH1-like protein (ASH1L) and similar proteins. This family includes SET domain-containing protein 2 (SETD2), nuclear SETD2 (NSD2) and ASH1-like protein (ASH1L), which function as histone-lysine N-methyltransferases. SETD2 specifically trimethylates 'Lys-36' of histone H3 (H3K36me3) using demethylated 'Lys-36' (H3K36me2) as substrate. NSD2 shows histone H3 'Lys-27' (H3K27me) methyltransferase activity. ASH1L specifically methylates 'Lys-36' of histone H3 (H3K36me). The family also includes Arabidopsis thaliana ASH1-related protein 3 (ASHR3) and similar proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.35e-71 | 378 | 698 | 69 | 373 | |
| 2.95e-71 | 378 | 700 | 59 | 371 | |
| 3.08e-70 | 378 | 698 | 53 | 361 | |
| 4.17e-70 | 379 | 705 | 53 | 367 | |
| 1.61e-69 | 378 | 698 | 53 | 361 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.27e-32 | 376 | 687 | 41 | 310 | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCU_A Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus] |
|
| 8.95e-22 | 563 | 738 | 39 | 217 | Chain A, 3-DEOXY-D-MANNO-OCTULOSONIC-ACID TRANSFERASE [Acinetobacter baumannii] |
|
| 2.62e-10 | 20 | 141 | 28 | 151 | The crystal structure of MLL3 SET domain [Homo sapiens] |
|
| 2.91e-10 | 20 | 141 | 32 | 155 | Crystal structure of the MLL3-Ash2L-RbBP5 complex [Homo sapiens],5F6K_E Crystal structure of the MLL3-Ash2L-RbBP5 complex [Homo sapiens] |
|
| 6.96e-10 | 20 | 141 | 78 | 201 | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.70e-53 | 378 | 707 | 57 | 383 | Probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial OS=Arabidopsis thaliana OX=3702 GN=KDTA PE=2 SV=1 |
|
| 2.79e-53 | 378 | 710 | 53 | 376 | 3-deoxy-D-manno-octulosonic acid transferase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=waaA PE=1 SV=1 |
|
| 2.50e-52 | 376 | 720 | 49 | 386 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli (strain K12) OX=83333 GN=waaA PE=1 SV=1 |
|
| 2.50e-52 | 376 | 720 | 49 | 386 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=waaA PE=3 SV=1 |
|
| 2.50e-52 | 376 | 720 | 49 | 386 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli O157:H7 OX=83334 GN=waaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
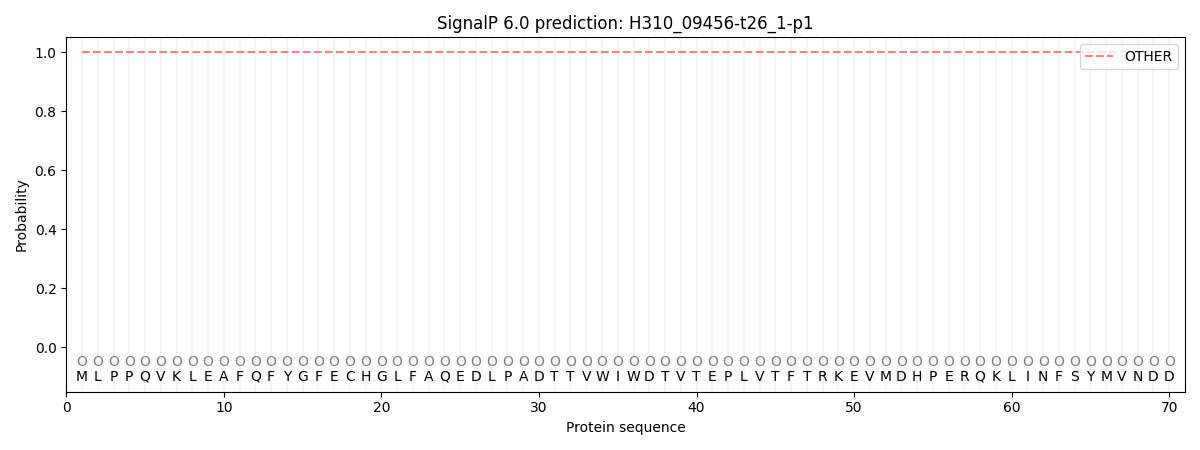
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000015 | 0.000000 |
