You are browsing environment: FUNGIDB
CAZyme Information: H310_07277-t26_5-p1
You are here: Home > Sequence: H310_07277-t26_5-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_07277-t26_5-p1 | |||||||||||
| CAZy Family | GH72 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 41 | 734 | 6.1e-75 | 0.5970744680851063 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225789 | LacZ | 6.85e-58 | 22 | 725 | 19 | 635 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 407625 | Ig_mannosidase | 5.50e-06 | 837 | 908 | 9 | 74 | Ig-fold domain. This Ig-like fold domain is found in mannosidase enzymes. |
| 395572 | Glyco_hydro_2 | 0.001 | 216 | 326 | 2 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.44e-228 | 19 | 892 | 28 | 844 | |
| 3.97e-185 | 42 | 895 | 80 | 966 | |
| 1.04e-119 | 43 | 908 | 41 | 868 | |
| 2.61e-117 | 41 | 877 | 46 | 850 | |
| 1.91e-114 | 22 | 909 | 29 | 874 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.09e-111 | 22 | 880 | 18 | 836 | mouse beta-mannosidase (MANBA) [Mus musculus],6DDU_A mouse beta-mannosidase bound to beta-D-mannose (MANBA) [Mus musculus] |
|
| 1.82e-84 | 42 | 892 | 29 | 819 | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VJX_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VL4_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VL4_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VMF_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VMF_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VO5_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VO5_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VOT_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VOT_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VQT_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron],2VQT_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron],2VR4_A Transition-state mimicry in mannoside hydrolysis: characterisation of twenty six inhibitors and insight into binding from linear free energy relationships and 3-D structure [Bacteroides thetaiotaomicron VPI-5482],2VR4_B Transition-state mimicry in mannoside hydrolysis: characterisation of twenty six inhibitors and insight into binding from linear free energy relationships and 3-D structure [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.88e-84 | 42 | 892 | 31 | 821 | Structure of a beta-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],2JE8_B Structure of a beta-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.92e-84 | 42 | 892 | 31 | 821 | Chain A, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP6_B Chain B, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP7_A Chain A, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP7_B Chain B, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482] |
|
| 4.85e-84 | 42 | 892 | 29 | 819 | Structure of the Michaelis complex of beta-mannosidase, Man2A, provides insight into the conformational itinerary of mannoside hydrolysis [Bacteroides thetaiotaomicron VPI-5482],2WBK_B Structure of the Michaelis complex of beta-mannosidase, Man2A, provides insight into the conformational itinerary of mannoside hydrolysis [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.60e-115 | 22 | 909 | 29 | 874 | Beta-mannosidase OS=Bos taurus OX=9913 GN=MANBA PE=1 SV=1 |
|
| 1.32e-113 | 22 | 909 | 29 | 874 | Beta-mannosidase OS=Capra hircus OX=9925 GN=MANBA PE=1 SV=1 |
|
| 7.21e-111 | 22 | 880 | 29 | 847 | Beta-mannosidase OS=Mus musculus OX=10090 GN=Manba PE=1 SV=1 |
|
| 1.02e-109 | 33 | 909 | 34 | 874 | Beta-mannosidase OS=Homo sapiens OX=9606 GN=MANBA PE=1 SV=3 |
|
| 1.07e-109 | 22 | 880 | 29 | 849 | Beta-mannosidase OS=Rattus norvegicus OX=10116 GN=Manba PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
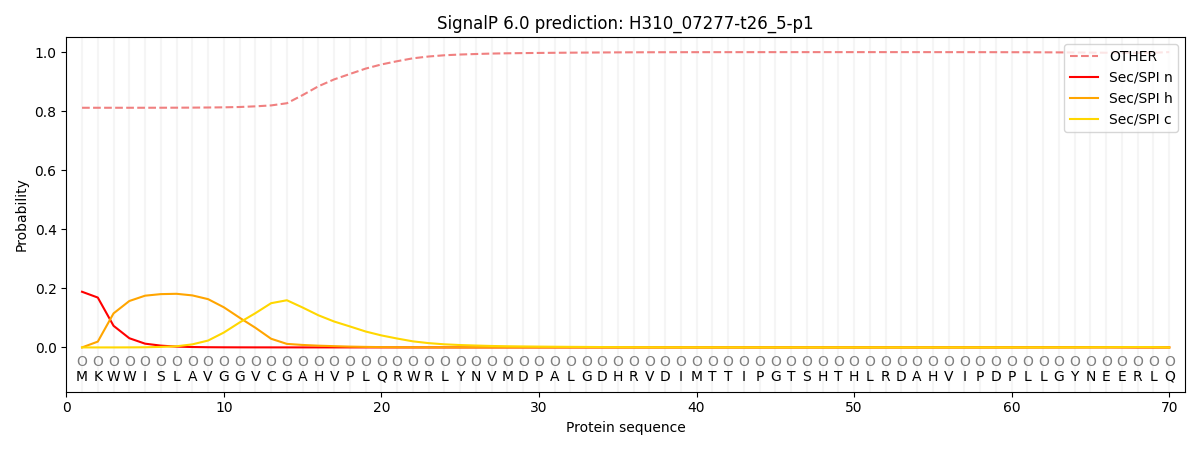
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.823612 | 0.176389 |
