You are browsing environment: FUNGIDB
CAZyme Information: H310_05438-t26_1-p1
You are here: Home > Sequence: H310_05438-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_05438-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 14 | 247 | 8.9e-69 | 0.9183673469387755 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 197593 | fCBD | 1.77e-07 | 306 | 337 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 197593 | fCBD | 3.91e-07 | 263 | 294 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 4.26e-06 | 263 | 289 | 2 | 28 | Fungal cellulose binding domain. |
| 395595 | CBM_1 | 1.44e-04 | 306 | 332 | 2 | 28 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.00e-195 | 1 | 307 | 1 | 307 | |
| 3.47e-140 | 1 | 307 | 1 | 336 | |
| 1.16e-70 | 15 | 295 | 14 | 271 | |
| 6.57e-70 | 15 | 295 | 14 | 271 | |
| 1.63e-65 | 24 | 301 | 21 | 268 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.59e-12 | 69 | 187 | 38 | 170 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.52e-08 | 260 | 300 | 18 | 58 | Endoglucanase gh5-1 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=gh5-1 PE=1 SV=1 |
|
| 2.91e-07 | 257 | 295 | 17 | 55 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
|
| 2.91e-07 | 257 | 295 | 17 | 55 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manF PE=3 SV=2 |
|
| 3.04e-07 | 254 | 300 | 15 | 60 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Sodiomyces alcalophilus OX=398408 PE=1 SV=1 |
|
| 7.95e-07 | 263 | 299 | 22 | 58 | Putative endoglucanase type F OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
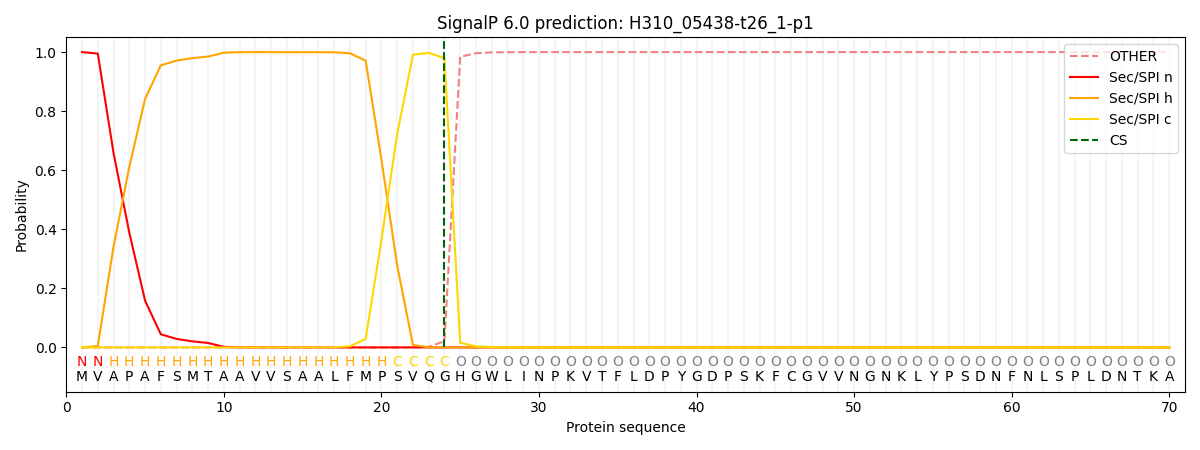
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000559 | 0.999407 | CS pos: 24-25. Pr: 0.9793 |
