You are browsing environment: FUNGIDB
CAZyme Information: H310_05432-t26_1-p1
You are here: Home > Sequence: H310_05432-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_05432-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 9 | 216 | 1.1e-71 | 0.8326530612244898 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 197593 | fCBD | 1.08e-08 | 267 | 298 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 197593 | fCBD | 5.15e-08 | 224 | 257 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 6.81e-06 | 267 | 294 | 2 | 29 | Fungal cellulose binding domain. |
| 395595 | CBM_1 | 1.20e-05 | 227 | 253 | 3 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.99e-210 | 1 | 268 | 1 | 268 | |
| 9.37e-102 | 10 | 266 | 12 | 279 | |
| 5.37e-101 | 10 | 266 | 12 | 279 | |
| 1.43e-78 | 9 | 259 | 11 | 297 | |
| 3.59e-74 | 21 | 257 | 22 | 261 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.48e-07 | 222 | 266 | 18 | 62 | Probable mannan endo-1,4-beta-mannosidase F OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manF PE=3 SV=2 |
|
| 1.49e-06 | 52 | 257 | 137 | 302 | Acetylxylan esterase OS=Hypocrea jecorina OX=51453 GN=axe1 PE=1 SV=1 |
|
| 1.60e-06 | 222 | 257 | 19 | 54 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
|
| 1.60e-06 | 222 | 257 | 19 | 54 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manF PE=3 SV=2 |
|
| 5.08e-06 | 228 | 257 | 435 | 464 | Endoglucanase 7a OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=eg7A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
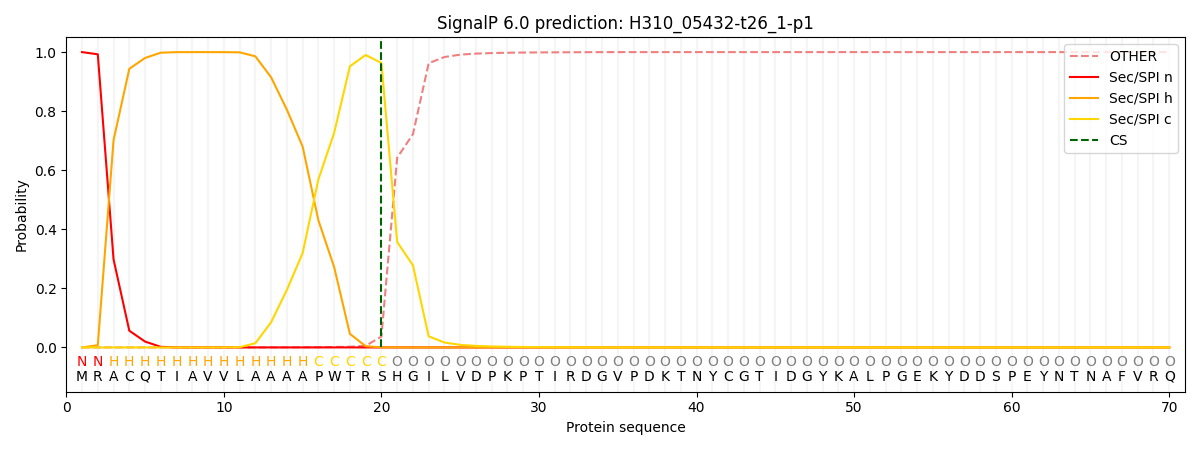
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000330 | 0.999649 | CS pos: 20-21. Pr: 0.9630 |
