You are browsing environment: FUNGIDB
CAZyme Information: H310_04069-t26_2-p1
You are here: Home > Sequence: H310_04069-t26_2-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
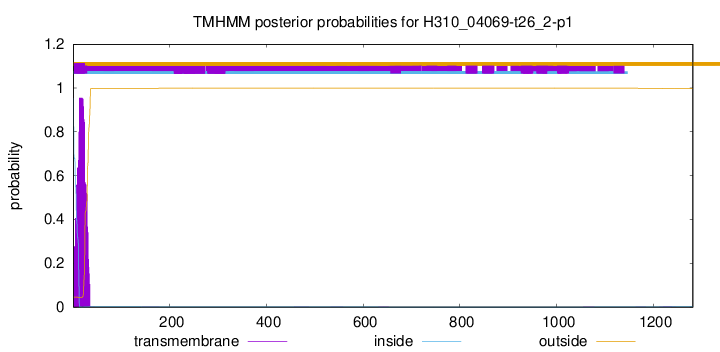
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_04069-t26_2-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 173 | 722 | 8.2e-102 | 0.64822695035461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 4.97e-92 | 114 | 723 | 57 | 616 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 404688 | Glyco_transf_41 | 1.17e-64 | 281 | 711 | 1 | 540 | Glycosyl transferase family 41. This family of glycosyltransferases includes O-linked beta-N-acetylglucosamine (O-GlcNAc) transferase, an enzyme which catalyzes the addition of O-GlcNAc to serine and threonine residues. In addition to its function as an O-GlcNAc transferase, human OGT also appears to proteolytically cleave the epigenetic cell-cycle regulator HCF-1. |
| 276809 | TPR | 3.86e-12 | 118 | 217 | 1 | 89 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 340831 | GT4_PimA-like | 1.67e-11 | 996 | 1274 | 7 | 327 | phosphatidyl-myo-inositol mannosyltransferase. This family is most closely related to the GT4 family of glycosyltransferases and named after PimA in Propionibacterium freudenreichii, which is involved in the biosynthesis of phosphatidyl-myo-inositol mannosides (PIM) which are early precursors in the biosynthesis of lipomannans (LM) and lipoarabinomannans (LAM), and catalyzes the addition of a mannosyl residue from GDP-D-mannose (GDP-Man) to the position 2 of the carrier lipid phosphatidyl-myo-inositol (PI) to generate a phosphatidyl-myo-inositol bearing an alpha-1,2-linked mannose residue (PIM1). Glycosyltransferases catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. The acceptor molecule can be a lipid, a protein, a heterocyclic compound, or another carbohydrate residue. This group of glycosyltransferases is most closely related to the previously defined glycosyltransferase family 1 (GT1). The members of this family may transfer UDP, ADP, GDP, or CMP linked sugars. The diverse enzymatic activities among members of this family reflect a wide range of biological functions. The protein structure available for this family has the GTB topology, one of the two protein topologies observed for nucleotide-sugar-dependent glycosyltransferases. GTB proteins have distinct N- and C- terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. The members of this family are found mainly in certain bacteria and archaea. |
| 276809 | TPR | 5.67e-11 | 165 | 261 | 3 | 96 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.01e-152 | 30 | 560 | 26 | 559 | |
| 5.82e-85 | 364 | 734 | 16 | 386 | |
| 1.45e-82 | 241 | 721 | 261 | 723 | |
| 3.58e-82 | 278 | 764 | 306 | 778 | |
| 4.22e-82 | 361 | 721 | 328 | 685 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.19e-53 | 175 | 655 | 35 | 495 | Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004],2VSN_B Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004] |
|
| 2.19e-53 | 175 | 655 | 35 | 495 | Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2JLB_B Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2VSY_A Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2VSY_B Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2XGM_A Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGM_B Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGO_A XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGO_B XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGS_A XcOGT in complex with C-UDP [Xanthomonas campestris],2XGS_B XcOGT in complex with C-UDP [Xanthomonas campestris] |
|
| 9.48e-48 | 281 | 721 | 158 | 705 | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation [Homo sapiens] |
|
| 1.58e-47 | 281 | 721 | 152 | 699 | The human O-GlcNAc transferase in complex with a thiol-linked bisubstrate inhibitor [Homo sapiens] |
|
| 1.60e-47 | 281 | 721 | 153 | 700 | The human O-GlcNAc transferase in complex with a bisubstrate inhibitor [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.93e-63 | 281 | 724 | 508 | 954 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC OS=Arabidopsis thaliana OX=3702 GN=SEC PE=1 SV=1 |
|
| 1.83e-46 | 281 | 721 | 476 | 1023 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Mus musculus OX=10090 GN=Ogt PE=1 SV=2 |
|
| 5.58e-46 | 281 | 721 | 476 | 1023 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Oryctolagus cuniculus OX=9986 GN=OGT PE=1 SV=2 |
|
| 7.36e-46 | 281 | 721 | 476 | 1023 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Homo sapiens OX=9606 GN=OGT PE=1 SV=3 |
|
| 1.70e-45 | 281 | 721 | 476 | 1023 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Sus scrofa OX=9823 GN=OGT PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
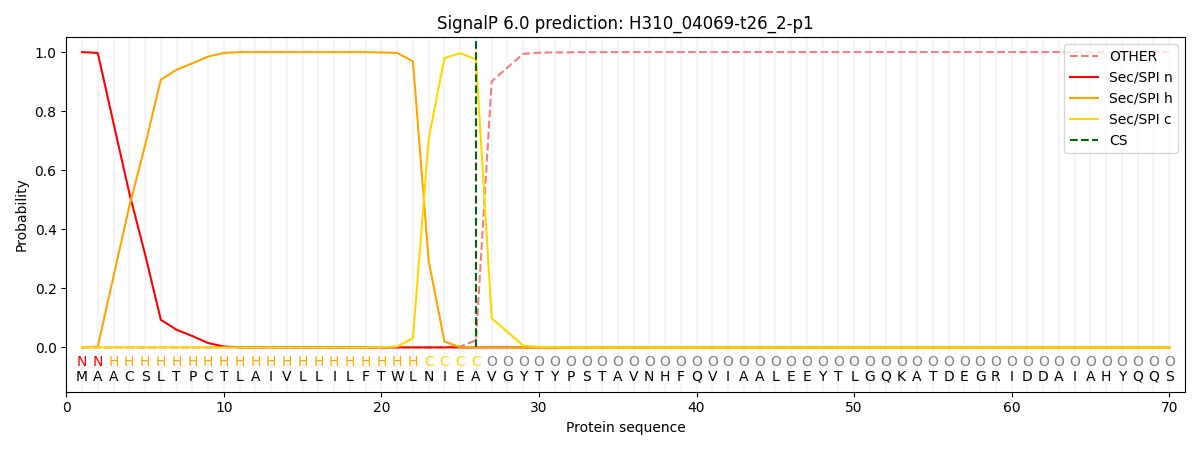
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000253 | 0.999743 | CS pos: 26-27. Pr: 0.9758 |