You are browsing environment: FUNGIDB
CAZyme Information: H310_01796-t26_1-p1
You are here: Home > Sequence: H310_01796-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_01796-t26_1-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.24:47 | 3.2.1.113:25 | 3.2.1.114:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH38 | 51 | 351 | 2e-74 | 0.9962825278810409 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 212121 | GH38N_AMII_LAM_like | 8.30e-140 | 50 | 314 | 1 | 278 | N-terminal catalytic domain of lysosomal alpha-mannosidase and similar proteins; glycoside hydrolase family 38 (GH38). The subfamily is represented by lysosomal alpha-mannosidase (LAM, Man2B1, EC 3.2.1.114), which is a broad specificity exoglycosidase hydrolyzing all known alpha 1,2-, alpha 1,3-, and alpha 1,6-mannosidic linkages from numerous high mannose type oligosaccharides. LAM is expressed in all tissues and in many species. In mammals, the absence of LAM can cause the autosomal recessive disease alpha-mannosidosis. LAM has an acidic pH optimum at 4.0-4.5. It is stimulated by zinc ion and is inhibited by cobalt ion and plant alkaloids, such as swainsonine (SW). LAM catalyzes hydrolysis by a double displacement mechanism in which a glycosyl-enzyme intermediate is formed and hydrolyzed via oxacarbenium ion-like transition states. A carboxylic acid in the active site acts as the catalytic nucleophile in the formation of the covalent intermediate while a second carboxylic acid acts as a general acid catalyst. The same residue is thought to assist in the hydrolysis (deglycosylation) step, this time acting as a general base. |
| 212095 | GH38N_AMII_euk | 1.96e-94 | 50 | 313 | 1 | 257 | N-terminal catalytic domain of eukaryotic class II alpha-mannosidases; glycoside hydrolase family 38 (GH38). The family corresponds to a group of eukaryotic class II alpha-mannosidases (AlphaMII), which contain Golgi alpha-mannosidases II (GMII), the major broad specificity lysosomal alpha-mannosidases (LAM, MAN2B1), the noval core-specific lysosomal alpha 1,6-mannosidases (Epman, MAN2B2), and similar proteins. GMII catalyzes the hydrolysis of the terminal both alpha-1,3-linked and alpha-1,6-linked mannoses from the high-mannose oligosaccharide GlcNAc(Man)5(GlcNAc)2 to yield GlcNAc(Man)3(GlcNAc)2 (GlcNAc, N-acetylglucosmine), which is the committed step of complex N-glycan synthesis. LAM is a broad specificity exoglycosidase hydrolyzing all known alpha 1,2-, alpha 1,3-, and alpha 1,6-mannosidic linkages from numerous high mannose type oligosaccharides. Different from LAM, Epman can efficiently cleave only the alpha 1,6-linked mannose residue from (Man)3GlcNAc, but not (Man)3(GlcNAc)2 or other larger high mannose oligosaccharides, in the core of N-linked glycans. Members in this family are retaining glycosyl hydrolases of family GH38 that employs a two-step mechanism involving the formation of a covalent glycosyl enzyme complex. Two carboxylic acids positioned within the active site act in concert: one as a catalytic nucleophile and the other as a general acid/base catalyst. |
| 178304 | PLN02701 | 2.47e-85 | 48 | 767 | 38 | 856 | alpha-mannosidase |
| 395852 | Glyco_hydro_38 | 4.39e-81 | 51 | 351 | 1 | 271 | Glycosyl hydrolases family 38 N-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. |
| 212120 | GH38N_AMII_GMII_SfManIII_like | 7.05e-74 | 49 | 362 | 1 | 340 | N-terminal catalytic domain of Golgi alpha-mannosidase II, Spodoptera frugiperda Sf9 alpha-mannosidase III, and similar proteins; glycoside hydrolase family 38 (GH38). This subfamily is represented by Golgi alpha-mannosidase II (GMII, also known as mannosyl-oligosaccharide 1,3- 1,6-alpha mannosidase, EC 3.2.1.114, Man2A1), a monomeric, membrane-anchored class II alpha-mannosidase existing in the Golgi apparatus of eukaryotes. GMII plays a key role in the N-glycosylation pathway. It catalyzes the hydrolysis of the terminal both alpha-1,3-linked and alpha-1,6-linked mannoses from the high-mannose oligosaccharide GlcNAc(Man)5(GlcNAc)2 to yield GlcNAc(Man)3(GlcNAc)2(GlcNAc, N-acetylglucosmine), which is the committed step of complex N-glycan synthesis. GMII is activated by zinc or cobalt ions and is strongly inhibited by swainsonine. Inhibition of GMII provides a route to block cancer-induced changes in cell surface oligosaccharide structures. GMII has a pH optimum of 5.5-6.0, which is intermediate between those of acidic (lysosomal alpha-mannosidase) and neutral (ER/cytosolic alpha-mannosidase) enzymes. GMII is a retaining glycosyl hydrolase of family GH38 that employs a two-step mechanism involving the formation of a covalent glycosyl enzyme complex; two carboxylic acids positioned within the active site act in concert: one as a catalytic nucleophile and the other as a general acid/base catalyst. This subfamily also includes human alpha-mannosidase 2x (MX, also known as mannosyl-oligosaccharide 1,3- 1,6-alpha mannosidase, EC 3.2.1.114, Man2A2). MX is enzymatically and functionally very similar to GMII, and is thought to also function in the N-glycosylation pathway. Also found in this subfamily is class II alpha-mannosidase encoded by Spodoptera frugiperda Sf9 cell. This alpha-mannosidase is an integral membrane glycoprotein localized in the Golgi apparatus. It shows high sequence homology with mammalian Golgi alpha-mannosidase II(GMII). It can hydrolyze p-nitrophenyl alpha-D-mannopyranoside (pNP-alpha-Man), and it is inhibited by swainsonine. However, the Sf9 enzyme is stimulated by cobalt and can hydrolyze (Man)5(GlcNAc)2 to (Man)3(GlcNAc)2, but it cannot hydrolyze GlcNAc(Man)5(GlcNAc)2, which is distinct from that of GMII. Thus, this enzyme has been designated as Sf9 alpha-mannosidase III (SfManIII). It probably functions in an alternate N-glycan processing pathway in Sf9 cells. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 26 | 950 | 19 | 950 | |
| 0.0 | 7 | 948 | 7 | 974 | |
| 5.24e-273 | 26 | 911 | 23 | 958 | |
| 6.48e-254 | 74 | 930 | 2 | 946 | |
| 9.25e-203 | 39 | 917 | 30 | 1000 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.72e-198 | 39 | 908 | 3 | 932 | Structure of GH 38 Jack Bean alpha-mannosidase [Canavalia ensiformis],6B9O_B Structure of GH 38 Jack Bean alpha-mannosidase [Canavalia ensiformis] |
|
| 9.72e-198 | 39 | 908 | 3 | 932 | Structure of GH 38 Jack Bean alpha-mannosidase in complex with a 36-valent iminosugar cluster inhibitor [Canavalia ensiformis],6B9P_B Structure of GH 38 Jack Bean alpha-mannosidase in complex with a 36-valent iminosugar cluster inhibitor [Canavalia ensiformis] |
|
| 6.08e-79 | 39 | 319 | 3 | 297 | The structure of the bovine lysosomal a-mannosidase suggests a novel mechanism for low pH activation [Bos taurus] |
|
| 1.56e-66 | 49 | 769 | 50 | 850 | GOLGI ALPHA-MANNOSIDASE II IN COMPLEX WITH SWAINSONINE [Drosophila melanogaster],1HXK_A Golgi Alpha-Mannosidase Ii In Complex With Deoxymannojirimicin [Drosophila melanogaster] |
|
| 1.81e-66 | 49 | 769 | 68 | 868 | Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RQZ_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RRH_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RRJ_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RRN_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RRU_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RRW_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RRX_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RRY_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster],6RS0_A Chain A, Alpha-mannosidase 2 [Drosophila melanogaster] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.90e-198 | 39 | 908 | 26 | 957 | Probable alpha-mannosidase At5g13980 OS=Arabidopsis thaliana OX=3702 GN=At5g13980 PE=2 SV=1 |
|
| 4.23e-188 | 39 | 908 | 3 | 932 | Alpha-mannosidase OS=Canavalia ensiformis OX=3823 PE=1 SV=1 |
|
| 8.78e-188 | 39 | 908 | 27 | 954 | Alpha-mannosidase At3g26720 OS=Arabidopsis thaliana OX=3702 GN=At3g26720 PE=1 SV=1 |
|
| 5.77e-178 | 39 | 908 | 36 | 985 | Probable alpha-mannosidase At5g66150 OS=Arabidopsis thaliana OX=3702 GN=At5g66150 PE=3 SV=1 |
|
| 1.23e-177 | 37 | 930 | 51 | 986 | Lysosomal alpha-mannosidase OS=Macaca fascicularis OX=9541 GN=MAN2B1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
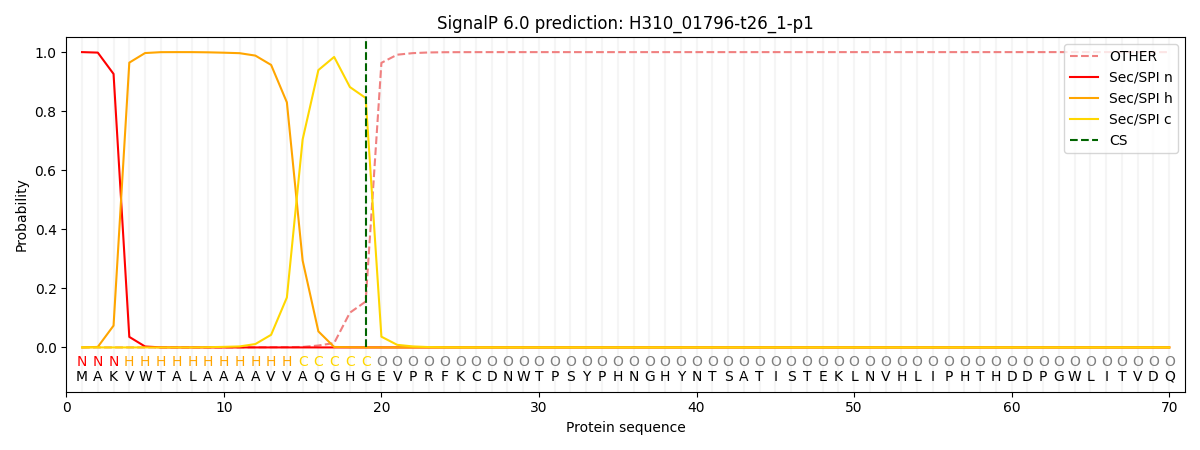
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000292 | 0.999660 | CS pos: 19-20. Pr: 0.8447 |
