You are browsing environment: FUNGIDB
CAZyme Information: H310_01558-t26_2-p1
You are here: Home > Sequence: H310_01558-t26_2-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_01558-t26_2-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.80:2 | 3.2.1.26:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM38 | 3087 | 3259 | 1.7e-22 | 0.9612403100775194 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 316802 | FG-GAP_2 | 6.62e-09 | 3043 | 3076 | 1 | 34 | FG-GAP repeat. |
| 316802 | FG-GAP_2 | 2.68e-07 | 2090 | 2137 | 1 | 49 | FG-GAP repeat. |
| 316802 | FG-GAP_2 | 1.71e-06 | 3315 | 3373 | 1 | 49 | FG-GAP repeat. |
| 197594 | Calx_beta | 1.83e-06 | 3446 | 3543 | 4 | 80 | Domains in Na-Ca exchangers and integrin-beta4. Domain in Na-Ca exchangers and integrin subunit beta4 (and some cyanobacterial proteins) |
| 316802 | FG-GAP_2 | 9.08e-06 | 2144 | 2191 | 1 | 49 | FG-GAP repeat. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.50e-29 | 3085 | 3275 | 68 | 222 | |
| 7.44e-29 | 492 | 1418 | 1867 | 2712 | |
| 4.98e-18 | 3087 | 3273 | 32 | 181 | |
| 1.88e-17 | 513 | 893 | 256 | 620 | |
| 3.19e-17 | 3087 | 3287 | 32 | 196 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.16e-09 | 3183 | 3275 | 367 | 458 | Putative glycosyl hydrolase ecdE OS=Aspergillus rugulosus OX=41736 GN=ecdE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
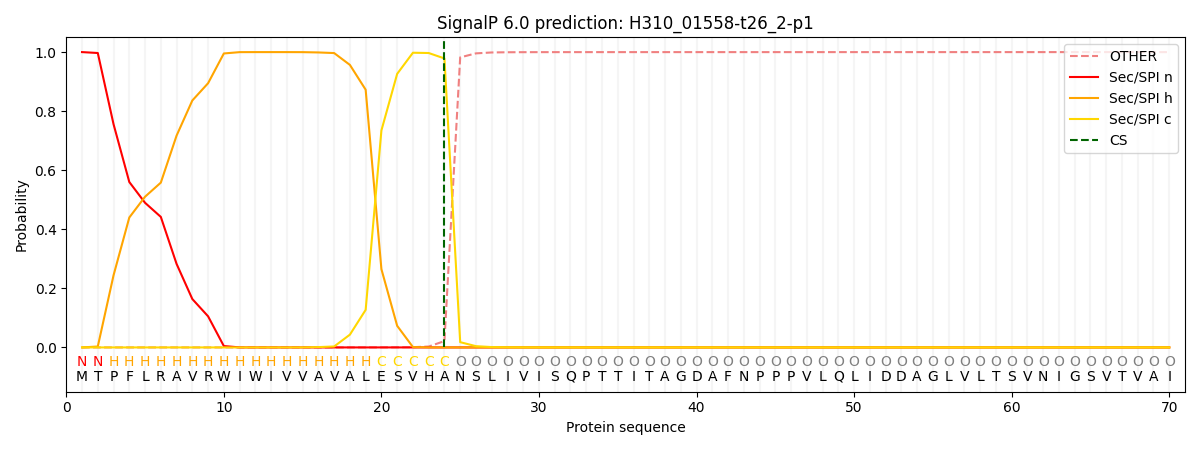
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000284 | 0.999688 | CS pos: 24-25. Pr: 0.9785 |
