You are browsing environment: FUNGIDB
CAZyme Information: H310_00388-t26_1-p1
You are here: Home > Sequence: H310_00388-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
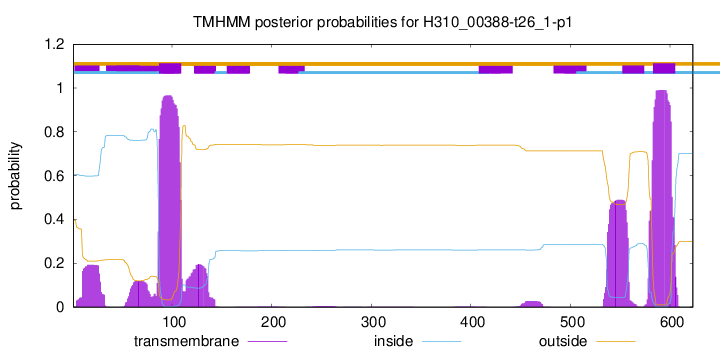
TMHMM annotations
Basic Information help
| Species | Aphanomyces invadans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans | |||||||||||
| CAZyme ID | H310_00388-t26_1-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 255 | 557 | 4.8e-25 | 0.725130890052356 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 1.02e-32 | 140 | 554 | 14 | 401 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 224732 | YjiC | 4.59e-23 | 132 | 562 | 7 | 403 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 278624 | UDPGT | 2.52e-13 | 148 | 596 | 21 | 480 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 223071 | egt | 5.79e-10 | 317 | 526 | 233 | 425 | ecdysteroid UDP-glucosyltransferase; Provisional |
| 215465 | PLN02863 | 1.46e-04 | 345 | 496 | 248 | 396 | UDP-glucoronosyl/UDP-glucosyl transferase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.67e-104 | 123 | 609 | 93 | 592 | |
| 2.89e-40 | 128 | 598 | 89 | 599 | |
| 1.03e-34 | 142 | 554 | 3 | 410 | |
| 6.19e-31 | 125 | 610 | 62 | 547 | |
| 7.93e-28 | 129 | 554 | 5 | 417 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.69e-07 | 455 | 554 | 312 | 411 | Structural analysis of EspG2 glycosyltransferase [Actinomadura verrucosospora],5DU2_B Structural analysis of EspG2 glycosyltransferase [Actinomadura verrucosospora] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.27e-14 | 234 | 583 | 141 | 489 | UDP-glucuronosyltransferase 3A1 OS=Bos taurus OX=9913 GN=UGT3A1 PE=2 SV=1 |
|
| 5.41e-14 | 222 | 540 | 135 | 446 | UDP-glucuronosyltransferase 1A4 OS=Oryctolagus cuniculus OX=9986 GN=Ugt1a4 PE=2 SV=1 |
|
| 2.85e-13 | 192 | 583 | 97 | 489 | UDP-glucuronosyltransferase 3A2 OS=Homo sapiens OX=9606 GN=UGT3A2 PE=2 SV=1 |
|
| 3.77e-13 | 275 | 583 | 179 | 489 | UDP-glucuronosyltransferase 3A1 OS=Mus musculus OX=10090 GN=Ugt3a1 PE=1 SV=1 |
|
| 6.60e-13 | 188 | 583 | 93 | 489 | UDP-glucuronosyltransferase 3A2 OS=Mus musculus OX=10090 GN=Ugt3a2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
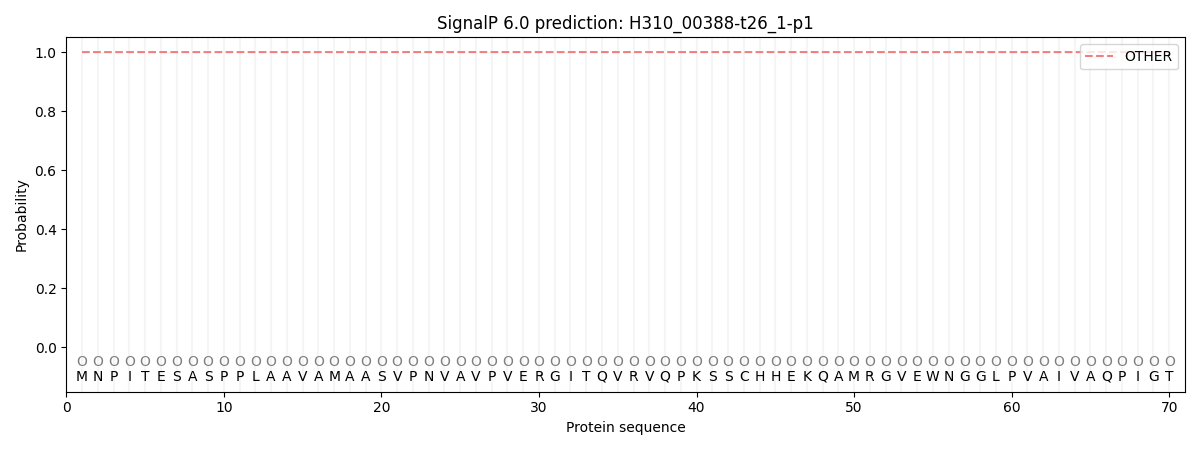
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000043 | 0.000005 |