You are browsing environment: FUNGIDB
CAZyme Information: H310_00119-t26_1-p1
Basic Information
help
| Species |
Aphanomyces invadans
|
| Lineage |
Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces invadans
|
| CAZyme ID |
H310_00119-t26_1-p1
|
| CAZy Family |
AA1 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 872 |
KI913952|CGC7 |
94302.77 |
6.5622 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AinvadansNJM9701 |
15416 |
N/A |
168 |
15248
|
|
| Gene Location |
No EC number prediction in H310_00119-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
262 |
635 |
1.6e-29 |
0.6698292220113852 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 404168
|
Beta_helix |
4.53e-05 |
518 |
642 |
1 |
126 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| 404168
|
Beta_helix |
0.002 |
519 |
573 |
94 |
148 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 7.50e-17 |
388 |
638 |
392 |
607 |
| 1.21e-15 |
393 |
638 |
437 |
649 |
| 3.72e-15 |
337 |
638 |
123 |
387 |
| 6.32e-14 |
399 |
635 |
238 |
462 |
| 1.90e-13 |
399 |
637 |
250 |
459 |
H310_00119-t26_1-p1 has no PDB hit.
H310_00119-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
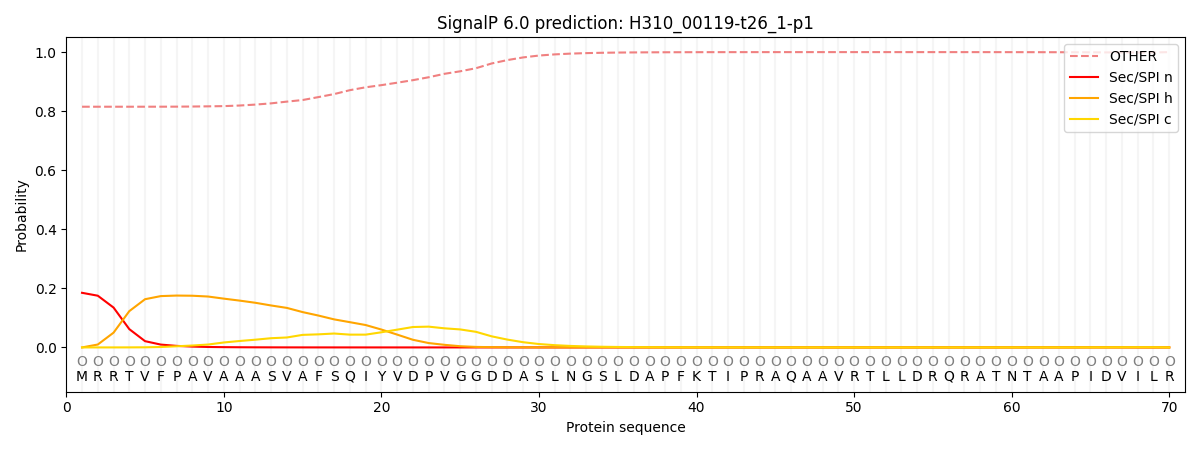
| Other |
SP_Sec_SPI |
CS Position |
| 0.827886 |
0.172100 |
|
There is no transmembrane helices in H310_00119-t26_1-p1.

