You are browsing environment: FUNGIDB
CAZyme Information: H257_15666-t26_1-p1
You are here: Home > Sequence: H257_15666-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_15666-t26_1-p1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 393 | 596 | 2.1e-18 | 0.7684887459807074 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 9.39e-11 | 309 | 592 | 38 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 227625 | Scw11 | 7.87e-09 | 635 | 901 | 44 | 305 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.20e-233 | 8 | 909 | 2 | 906 | |
| 7.22e-163 | 23 | 564 | 20 | 559 | |
| 4.44e-96 | 301 | 592 | 8 | 304 | |
| 2.14e-89 | 304 | 605 | 10 | 319 | |
| 4.42e-88 | 311 | 611 | 16 | 318 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.40e-07 | 394 | 598 | 145 | 382 | Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_B Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_C Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_D Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_E Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_F Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
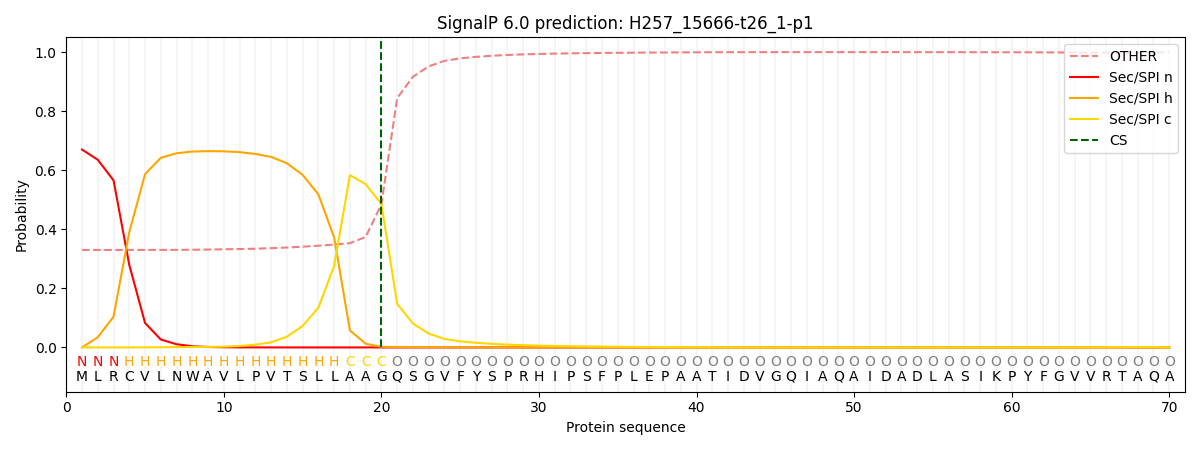
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.346315 | 0.653658 | CS pos: 20-21. Pr: 0.4855 |
