You are browsing environment: FUNGIDB
CAZyme Information: H257_14650-t26_1-p1
You are here: Home > Sequence: H257_14650-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_14650-t26_1-p1 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.53:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA15 | 24 | 208 | 9.8e-52 | 0.9948186528497409 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397269 | LPMO_10 | 3.57e-10 | 102 | 208 | 89 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 516 | 1 | 516 | |
| 1.29e-176 | 1 | 397 | 1 | 398 | |
| 2.09e-163 | 1 | 311 | 1 | 286 | |
| 3.11e-162 | 7 | 516 | 7 | 463 | |
| 8.76e-138 | 11 | 311 | 11 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.66e-23 | 24 | 211 | 1 | 193 | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State [Thermobia domestica] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
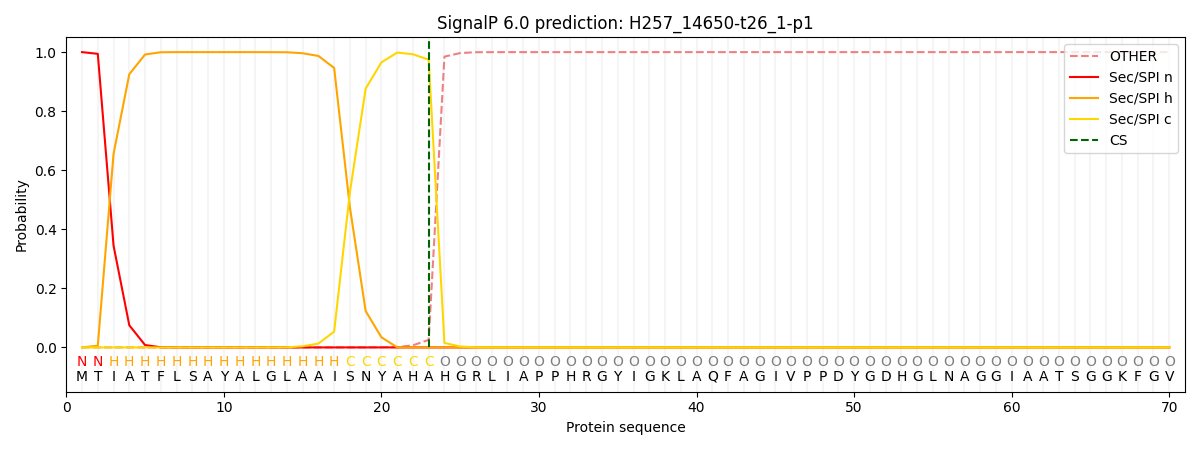
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000255 | 0.999689 | CS pos: 23-24. Pr: 0.9745 |
