You are browsing environment: FUNGIDB
CAZyme Information: H257_12919-t26_8-p1
You are here: Home > Sequence: H257_12919-t26_8-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_12919-t26_8-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | trehalose-phosphatase, variant 6 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT20 | 22 | 289 | 5.7e-54 | 0.5621052631578948 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 177855 | PLN02205 | 1.24e-76 | 22 | 596 | 263 | 847 | alpha,alpha-trehalose-phosphate synthase [UDP-forming] |
| 184712 | PRK14501 | 1.38e-75 | 22 | 595 | 195 | 722 | putative bifunctional trehalose-6-phosphate synthase/HAD hydrolase subfamily IIB; Provisional |
| 215555 | PLN03063 | 2.70e-51 | 22 | 604 | 209 | 791 | alpha,alpha-trehalose-phosphate synthase (UDP-forming); Provisional |
| 215556 | PLN03064 | 1.10e-48 | 22 | 520 | 293 | 788 | alpha,alpha-trehalose-phosphate synthase (UDP-forming); Provisional |
| 340820 | GT20_TPS | 7.95e-46 | 22 | 288 | 196 | 462 | trehalose-6-phosphate synthase. Trehalose-6-Phosphate Synthase (TPS, EC 2.4.1.15) is a glycosyltransferase that catalyses the synthesis of alpha,alpha-1,1-trehalose-6-phosphate from glucose-6-phosphate using a UDP-glucose donor. It is a key enzyme in the trehalose synthesis pathway. Trehalose is a nonreducing disaccharide present in a wide variety of organisms and may serve as a source of energy and carbon. It is characterized most notably in insect, plant, and microbial cells. Its production is often associated with a variety of stress conditions, including desiccation, dehydration, heat, cold, and oxidation. This family represents the catalytic domain of the TPS. Some members of this domain family coexist with a C-terminal trehalose phosphatase domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.90e-174 | 22 | 616 | 246 | 849 | |
| 8.68e-151 | 46 | 615 | 266 | 845 | |
| 4.90e-82 | 22 | 613 | 504 | 1135 | |
| 4.42e-81 | 14 | 613 | 469 | 1093 | |
| 2.18e-75 | 14 | 593 | 535 | 1162 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.71e-31 | 303 | 580 | 3 | 251 | Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 2 [Aspergillus fumigatus Af293],5DXO_A Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 3 [Aspergillus fumigatus Af293],5DXO_B Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 3 [Aspergillus fumigatus Af293] |
|
| 5.53e-30 | 303 | 580 | 3 | 251 | Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 1 [Aspergillus fumigatus Af293] |
|
| 2.97e-26 | 302 | 557 | 8 | 246 | Structure of C. albicans Trehalose-6-phosphate phosphatase C-terminal domain [Candida albicans SC5314],5DXI_B Structure of C. albicans Trehalose-6-phosphate phosphatase C-terminal domain [Candida albicans SC5314] |
|
| 2.39e-25 | 316 | 554 | 15 | 222 | Chain A, trehalose-6-phosphate phosphatase [Cryptococcus neoformans] |
|
| 6.49e-25 | 14 | 288 | 193 | 467 | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP-glucose [Candida albicans SC5314],5HUT_B Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP-glucose [Candida albicans SC5314],5HUU_A Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and glucose-6-phosphate [Candida albicans SC5314],5HUU_B Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and glucose-6-phosphate [Candida albicans SC5314],5HVL_A Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A [Candida albicans SC5314],5HVL_B Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A [Candida albicans SC5314] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.10e-70 | 22 | 599 | 251 | 834 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 11 OS=Arabidopsis thaliana OX=3702 GN=TPS11 PE=2 SV=1 |
|
| 2.98e-68 | 22 | 601 | 268 | 858 | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 OS=Arabidopsis thaliana OX=3702 GN=TPS6 PE=1 SV=2 |
|
| 3.41e-67 | 22 | 599 | 258 | 836 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 OS=Arabidopsis thaliana OX=3702 GN=TPS7 PE=1 SV=1 |
|
| 3.58e-66 | 22 | 608 | 260 | 851 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 10 OS=Arabidopsis thaliana OX=3702 GN=TPS10 PE=2 SV=1 |
|
| 1.37e-65 | 22 | 603 | 260 | 846 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 OS=Arabidopsis thaliana OX=3702 GN=TPS9 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
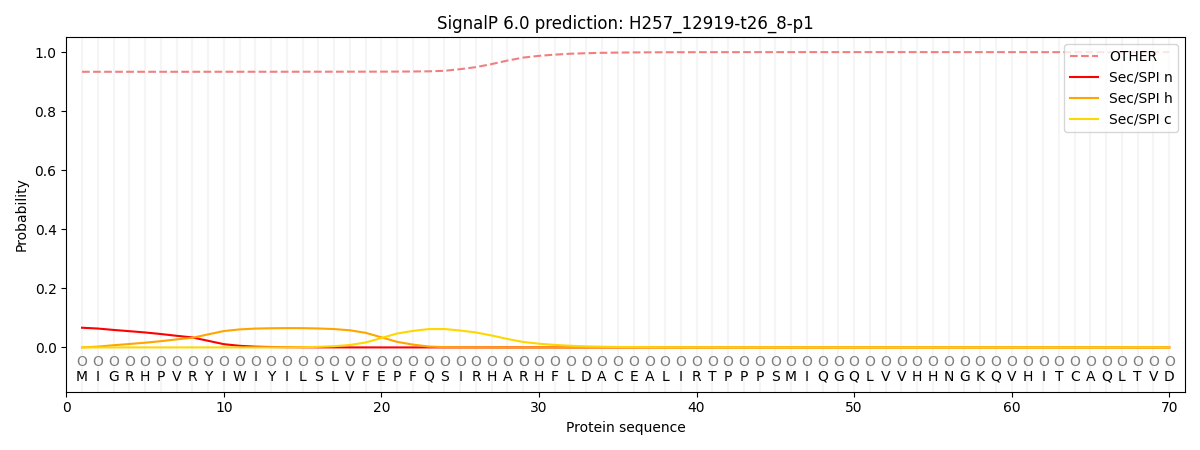
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.937275 | 0.062723 |
