You are browsing environment: FUNGIDB
CAZyme Information: H257_11264-t26_1-p1
You are here: Home > Sequence: H257_11264-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
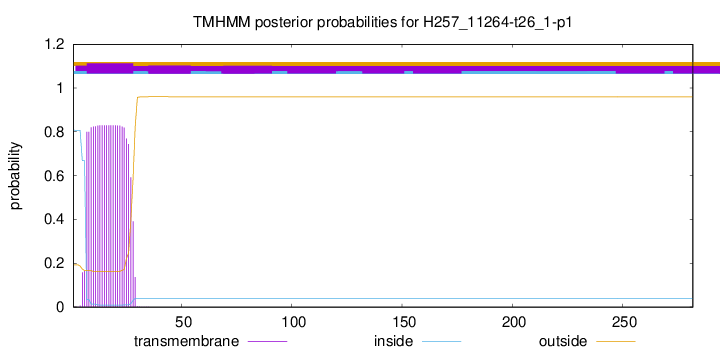
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_11264-t26_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 11 | 234 | 4.6e-76 | 0.9020408163265307 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 197593 | fCBD | 3.38e-08 | 238 | 269 | 2 | 33 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 6.94e-08 | 238 | 266 | 1 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.67e-211 | 1 | 282 | 1 | 282 | |
| 1.37e-210 | 1 | 282 | 1 | 282 | |
| 2.04e-95 | 21 | 279 | 18 | 266 | |
| 1.57e-78 | 21 | 271 | 19 | 262 | |
| 1.90e-76 | 20 | 271 | 19 | 296 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.92e-17 | 70 | 190 | 44 | 171 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.36e-09 | 238 | 274 | 22 | 58 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manF PE=3 SV=1 |
|
| 2.70e-08 | 239 | 271 | 21 | 53 | Endoglucanase 3 OS=Humicola insolens OX=34413 GN=CMC3 PE=1 SV=1 |
|
| 7.05e-08 | 235 | 279 | 19 | 63 | Mannan endo-1,4-beta-mannosidase F OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=manF PE=3 SV=1 |
|
| 2.35e-07 | 235 | 279 | 19 | 63 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manF PE=3 SV=2 |
|
| 2.35e-07 | 235 | 279 | 19 | 63 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000268 | 0.999687 | CS pos: 23-24. Pr: 0.9809 |