You are browsing environment: FUNGIDB
CAZyme Information: H257_10110-t26_1-p1
You are here: Home > Sequence: H257_10110-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
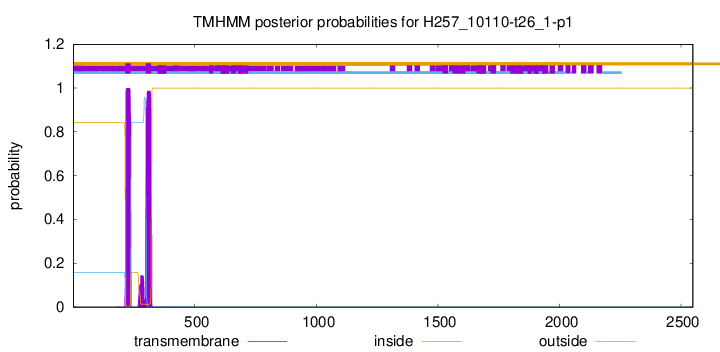
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_10110-t26_1-p1 | |||||||||||
| CAZy Family | GT76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.12:3 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350188 | SF2_C_FANCM_Hef | 8.80e-50 | 973 | 1109 | 9 | 142 | C-terminal helicase domain of Fanconi anemia group M family helicases. Fanconi anemia group M (FANCM) protein is a DNA-dependent ATPase component of the Fanconi anemia (FA) core complex. It is required for the normal activation of the FA pathway, leading to monoubiquitination of the FANCI-FANCD2 complex in response to DNA damage, cellular resistance to DNA cross-linking drugs, and prevention of chromosomal breakage. Hef (helicase-associated endonuclease fork-structure) belongs to the XPF/MUS81/FANCM family of endonucleases and is involved in stalled replication fork repair. FANCM and Hef are DEAD-like helicases belonging to superfamily (SF)2, a diverse family of proteins involved in ATP-dependent RNA or DNA unwinding. Similar to SF1 helicases, SF2 helicases do not form toroidal structures like SF3-6 helicases. Their helicase core consists of two similar protein domains that resemble the fold of the recombination protein RecA. This model describes the C-terminal domain, also called HelicC. |
| 224036 | MPH1 | 1.59e-49 | 906 | 1161 | 281 | 530 | ERCC4-related helicase [Replication, recombination and repair]. |
| 133043 | CESA_CelA_like | 2.77e-44 | 346 | 786 | 1 | 234 | CESA_CelA_like are involved in the elongation of the glucan chain of cellulose. Family of proteins related to Agrobacterium tumefaciens CelA and Gluconacetobacter xylinus BscA. These proteins are involved in the elongation of the glucan chain of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues. They are putative catalytic subunit of cellulose synthase, which is a glycosyltransferase using UDP-glucose as the substrate. The catalytic subunit is an integral membrane protein with 6 transmembrane segments and it is postulated that the protein is anchored in the membrane at the N-terminal end. |
| 237496 | PRK13766 | 1.50e-35 | 906 | 1123 | 282 | 500 | Hef nuclease; Provisional |
| 227470 | COG5141 | 7.37e-28 | 1768 | 1920 | 188 | 338 | PHD zinc finger-containing protein [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 8 | 819 | 11 | 817 | |
| 0.0 | 11 | 819 | 8 | 809 | |
| 0.0 | 7 | 819 | 11 | 810 | |
| 0.0 | 7 | 819 | 11 | 812 | |
| 0.0 | 7 | 819 | 11 | 810 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.63e-31 | 1776 | 1935 | 3 | 165 | X-ray crystal structure of BRPF1 PZP domain [Homo sapiens] |
|
| 9.28e-31 | 1776 | 1935 | 23 | 185 | Crystal structure of human BRPF1 PZP bound to histone H3 tail [Homo sapiens] |
|
| 1.17e-26 | 894 | 1116 | 264 | 478 | Chain A, ATP-dependent RNA helicase, putative [Pyrococcus furiosus DSM 3638],1WP9_B Chain B, ATP-dependent RNA helicase, putative [Pyrococcus furiosus DSM 3638],1WP9_C Chain C, ATP-dependent RNA helicase, putative [Pyrococcus furiosus DSM 3638],1WP9_D Chain D, ATP-dependent RNA helicase, putative [Pyrococcus furiosus DSM 3638],1WP9_E Chain E, ATP-dependent RNA helicase, putative [Pyrococcus furiosus DSM 3638],1WP9_F Chain F, ATP-dependent RNA helicase, putative [Pyrococcus furiosus DSM 3638] |
|
| 3.78e-26 | 1776 | 1942 | 23 | 197 | Chain A, Histone H3.1,Protein AF-10 [Homo sapiens] |
|
| 3.99e-26 | 1776 | 1942 | 25 | 199 | Crystal structure of PZP domain of human AF10 protein [Homo sapiens],5DAH_A Crystal structure of PZP domain of human AF10 protein fused with Histone H3 peptide [Homo sapiens],5DAH_B Crystal structure of PZP domain of human AF10 protein fused with Histone H3 peptide [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.83e-47 | 866 | 1157 | 311 | 635 | Fanconi anemia group M protein OS=Gallus gallus OX=9031 GN=FANCM PE=1 SV=1 |
|
| 3.02e-46 | 825 | 1206 | 288 | 695 | Fanconi anemia group M protein OS=Homo sapiens OX=9606 GN=FANCM PE=1 SV=2 |
|
| 2.24e-44 | 825 | 1157 | 276 | 622 | Fanconi anemia group M protein homolog OS=Mus musculus OX=10090 GN=Fancm PE=1 SV=3 |
|
| 3.92e-41 | 872 | 1165 | 388 | 686 | ATP-dependent DNA helicase mph1 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=mph1 PE=3 SV=1 |
|
| 2.90e-40 | 819 | 1165 | 480 | 811 | ATP-dependent DNA helicase MPH1 OS=Botryotinia fuckeliana (strain B05.10) OX=332648 GN=mph1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
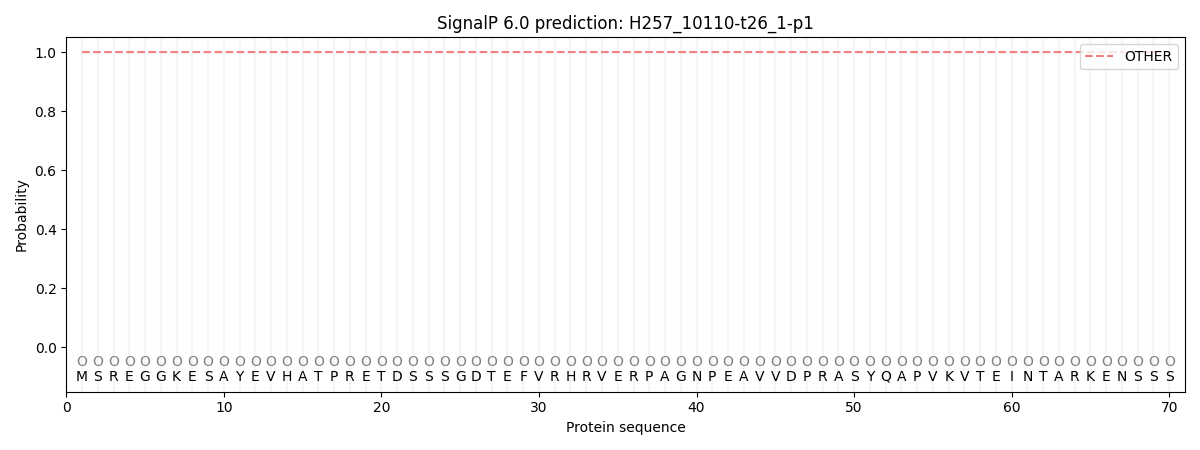
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000057 | 0.000000 |