You are browsing environment: FUNGIDB
CAZyme Information: H257_08158-t26_2-p1
You are here: Home > Sequence: H257_08158-t26_2-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_08158-t26_2-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 2 | 342 | 1.1e-83 | 0.8431876606683805 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215437 | PLN02816 | 3.23e-85 | 1 | 422 | 98 | 533 | mannosyltransferase |
| 281842 | Glyco_transf_22 | 1.56e-62 | 2 | 342 | 61 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.13e-89 | 2 | 422 | 103 | 531 | |
| 1.02e-87 | 1 | 422 | 17 | 452 | |
| 9.13e-87 | 1 | 422 | 98 | 533 | |
| 9.13e-87 | 1 | 422 | 98 | 533 | |
| 9.13e-87 | 1 | 422 | 98 | 533 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.62e-87 | 1 | 422 | 98 | 533 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
|
| 4.22e-76 | 1 | 415 | 102 | 508 | GPI mannosyltransferase 3 OS=Danio rerio OX=7955 GN=pigb PE=2 SV=1 |
|
| 1.28e-75 | 2 | 391 | 121 | 502 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
|
| 3.65e-75 | 2 | 437 | 111 | 534 | GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1 |
|
| 2.86e-74 | 2 | 415 | 110 | 515 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000031 | 0.000005 |
TMHMM Annotations download full data without filtering help
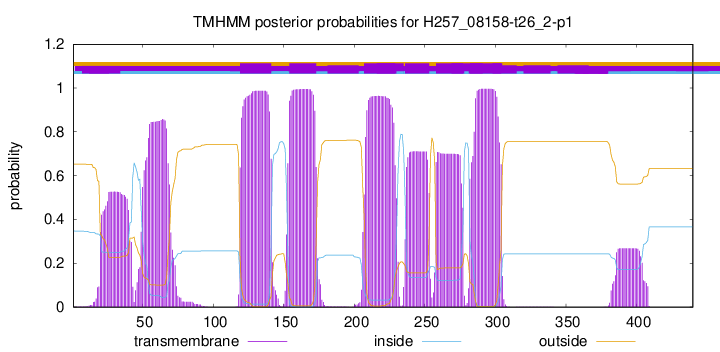
| Start | End |
|---|---|
| 119 | 141 |
| 154 | 173 |
| 207 | 229 |
| 236 | 253 |
| 258 | 277 |
| 282 | 304 |
