You are browsing environment: FUNGIDB
CAZyme Information: H257_07745-t26_1-p1
You are here: Home > Sequence: H257_07745-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_07745-t26_1-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 1074 | 1795 | 4.8e-272 | 0.9566982408660352 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 1074 | 1800 | 3 | 765 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 2.83e-28 | 226 | 315 | 8 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 8 | 1964 | 20 | 2001 | |
| 0.0 | 4 | 1964 | 16 | 2005 | |
| 0.0 | 9 | 1963 | 21 | 2002 | |
| 1.01e-266 | 134 | 1820 | 91 | 1612 | |
| 5.44e-261 | 134 | 1957 | 98 | 1752 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.35e-246 | 125 | 1960 | 239 | 1916 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 4.43e-240 | 118 | 1961 | 225 | 1938 | Callose synthase 1 OS=Arabidopsis thaliana OX=3702 GN=CALS1 PE=1 SV=2 |
|
| 6.85e-240 | 118 | 1961 | 229 | 1943 | Callose synthase 3 OS=Arabidopsis thaliana OX=3702 GN=CALS3 PE=3 SV=3 |
|
| 1.69e-238 | 125 | 1963 | 250 | 1884 | Callose synthase 9 OS=Arabidopsis thaliana OX=3702 GN=CALS9 PE=2 SV=2 |
|
| 2.30e-237 | 118 | 1961 | 225 | 1938 | Callose synthase 2 OS=Arabidopsis thaliana OX=3702 GN=CALS2 PE=2 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
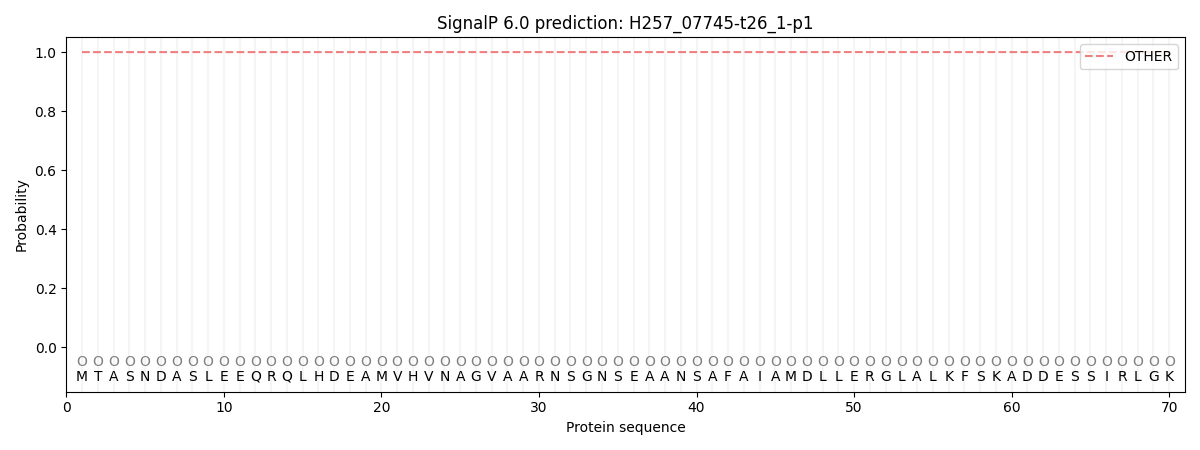
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000034 | 0.000001 |
TMHMM Annotations download full data without filtering help
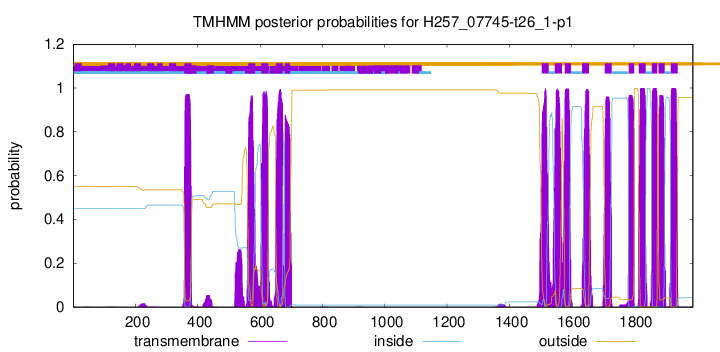
| Start | End |
|---|---|
| 1879 | 1898 |
| 1918 | 1940 |
| 357 | 379 |
| 562 | 584 |
| 604 | 626 |
| 650 | 672 |
| 679 | 696 |
| 1504 | 1526 |
| 1546 | 1568 |
| 1578 | 1597 |
| 1633 | 1655 |
| 1706 | 1728 |
| 1783 | 1800 |
| 1815 | 1837 |
| 1857 | 1874 |
