You are browsing environment: FUNGIDB
CAZyme Information: H257_06586-t26_1-p1
Basic Information
help
| Species |
Aphanomyces astaci
|
| Lineage |
Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci
|
| CAZyme ID |
H257_06586-t26_1-p1
|
| CAZy Family |
GH18 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 899 |
KI913126|CGC6 |
97159.30 |
5.8412 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AastaciAPO3 |
19584 |
N/A |
465 |
19119
|
|
| Gene Location |
No EC number prediction in H257_06586-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
392 |
662 |
1.1e-26 |
0.5028462998102466 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 404168
|
Beta_helix |
3.13e-04 |
546 |
675 |
25 |
134 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 5.41e-16 |
326 |
657 |
309 |
599 |
| 6.62e-15 |
424 |
657 |
441 |
641 |
| 1.58e-14 |
395 |
657 |
157 |
379 |
| 5.46e-14 |
424 |
673 |
235 |
498 |
| 6.53e-14 |
426 |
723 |
246 |
547 |
H257_06586-t26_1-p1 has no PDB hit.
H257_06586-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
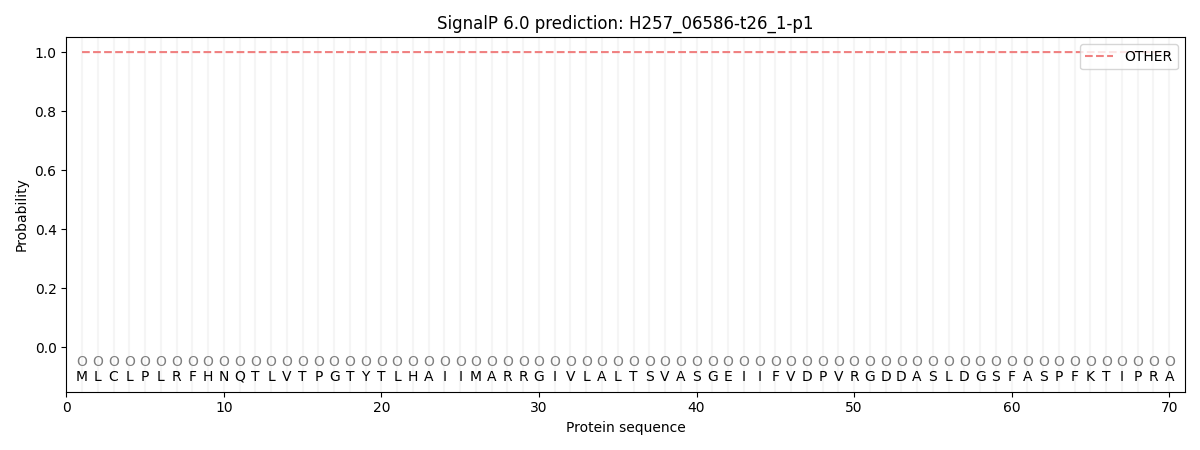
| Other |
SP_Sec_SPI |
CS Position |
| 1.000037 |
0.000009 |
|
There is no transmembrane helices in H257_06586-t26_1-p1.

