You are browsing environment: FUNGIDB
CAZyme Information: H257_05174-t26_1-p1
You are here: Home > Sequence: H257_05174-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_05174-t26_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.37:92 | 3.2.1.55:39 | 3.2.1.21:1 | 3.2.1.6:1 | 3.2.1.73:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 78 | 317 | 3.1e-68 | 0.9629629629629629 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178629 | PLN03080 | 0.0 | 6 | 735 | 10 | 778 | Probable beta-xylosidase; Provisional |
| 185053 | PRK15098 | 2.08e-69 | 109 | 702 | 118 | 728 | beta-glucosidase BglX. |
| 224389 | BglX | 6.02e-60 | 69 | 418 | 49 | 365 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| 396478 | Glyco_hydro_3_C | 8.57e-45 | 387 | 618 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| 395747 | Glyco_hydro_3 | 6.78e-34 | 108 | 315 | 85 | 281 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.75e-170 | 31 | 735 | 43 | 775 | |
| 3.96e-168 | 28 | 738 | 28 | 771 | |
| 3.96e-168 | 28 | 726 | 28 | 759 | |
| 4.67e-167 | 28 | 735 | 33 | 764 | |
| 4.67e-167 | 28 | 735 | 33 | 764 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.18e-138 | 28 | 725 | 5 | 717 | Chain A, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
|
| 4.18e-138 | 28 | 725 | 5 | 717 | Chain A, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
|
| 5.45e-118 | 28 | 732 | 32 | 746 | GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
|
| 8.80e-107 | 28 | 744 | 32 | 761 | Chain A, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
|
| 9.02e-107 | 28 | 744 | 32 | 761 | Chain A, BETA-XYLOSIDASE [Trichoderma reesei],5AE6_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.49e-164 | 12 | 735 | 16 | 759 | Probable beta-D-xylosidase 2 OS=Arabidopsis thaliana OX=3702 GN=BXL2 PE=2 SV=1 |
|
| 2.60e-164 | 21 | 729 | 22 | 761 | Probable beta-D-xylosidase 5 OS=Arabidopsis thaliana OX=3702 GN=BXL5 PE=2 SV=2 |
|
| 9.00e-163 | 38 | 734 | 49 | 771 | Beta-D-xylosidase 3 OS=Arabidopsis thaliana OX=3702 GN=BXL3 PE=1 SV=1 |
|
| 1.03e-161 | 7 | 734 | 20 | 772 | Beta-xylosidase/alpha-L-arabinofuranosidase 2 OS=Medicago sativa subsp. varia OX=36902 GN=Xyl2 PE=2 SV=1 |
|
| 3.37e-159 | 28 | 735 | 46 | 783 | Beta-D-xylosidase 4 OS=Arabidopsis thaliana OX=3702 GN=BXL4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
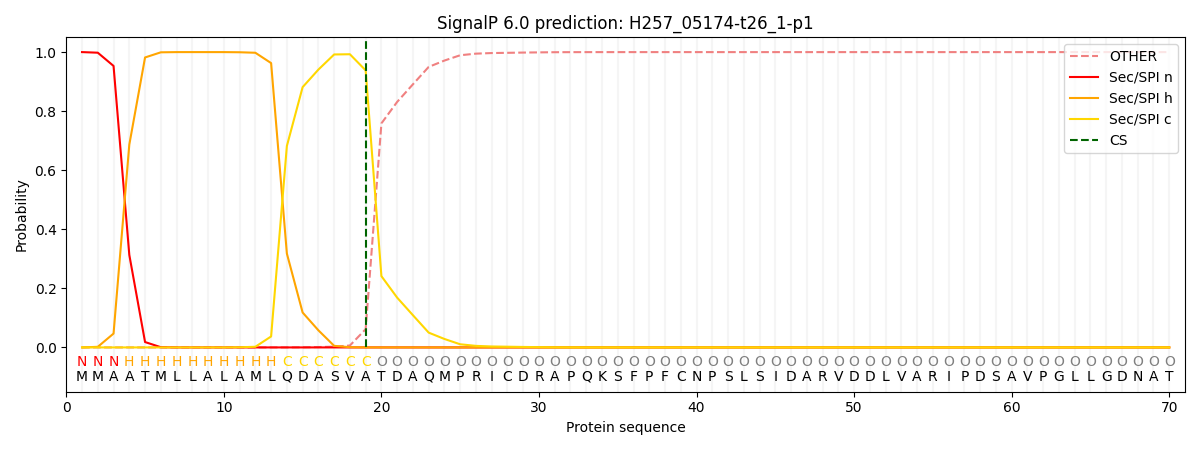
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000282 | 0.999704 | CS pos: 19-20. Pr: 0.9382 |
